Simulation in RsNLME
simulation.Rmd
Overview
The purpose of this vignette is to demonstrate how to edit a built-in model and then simulate it.
The model demonstrated is a two-compartment model with an IV bolus. It involves both plasma and urine compartments with the urine compartment reset to zero right after each observation.
Create the model
It is assumed that directory with NLME executables is given as an
environment variable (INSTALLDIR), which can be checked
with the following commands:
# Check the environment variable
Sys.getenv("INSTALLDIR")First we will create the simulation input dataset.
# loading the package
library(data.table)
dt_SimInputData <- data.table(ID = c(1, 2), Time = 0, Dose = c(10, 20))Next, we will define the model and the associate column mappings:
- Create a basic two-compartment PK model with IV bolus and define the associated column mappings.
- Change initial values for fixed effects tvV, tvV2, and tvCl2, to 5, 3, 0.5, respectively.
- Change the covariance matrix of random effects to be a diagonal matrix with all the diagonal elements being 0.1.
- Change the standard deviation of residual error variable corresponding to observed variable A0Obs to 0.2.
library(Certara.RsNLME)
library(ggplot2)
library(dplyr)
ModelName <- "TwoCpt_IVBolus_1stOrderElim_PlasmaUrineObs"
model <-
pkmodel(
numCompartments = 2,
hasEliminationComp = TRUE,
isClosedForm = FALSE,
data = dt_SimInputData,
ID = "ID",
Time = "Time",
A1 = "Dose",
modelName = ModelName
) %>%
fixedEffect(effect = c("tvV", "tvV2", "tvCl2"),
value = c(5, 3, 0.5)) %>%
randomEffect(effect = c("nV", "nCl", "nV2", "nCl2"),
value = rep(0.1, 4)) %>%
residualError(predName = "A0", SD = 0.2)Let’s edit the model using the editModel function.
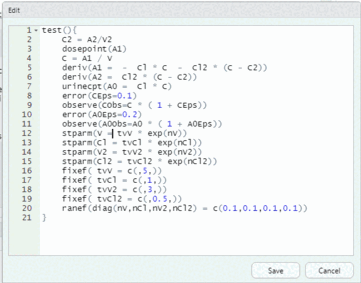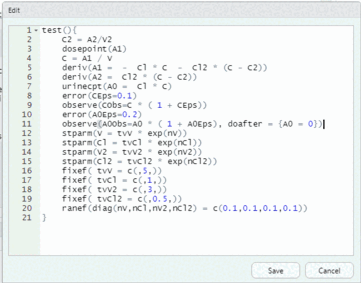
model <- editModel(model)To reset the urine compartment to 0 right after each observation, add “, doafter = {A0 = 0}” inside the observe statement for A0Obs; that is, change the following statement:
observe(A0Obs=A0 * ( 1 + A0Eps))to
observe(A0Obs=A0 * ( 1 + A0Eps), doafter = {A0 = 0})and then click the “Save” button to update the model.
Model Simulation
We will define simulation tables for plasma and urine observations, set the number of replicates to be 50, and change the starting seed to 1:
## Define simulation tables for plasma observations
SimTableCObs <- tableParams(
name = "SimTableCObs.csv",
timesList = c(0, 0.5, 1, 2, 4, 8, 12, 16, 20, 24),
variablesList = c("C", "CObs"),
forSimulation = TRUE
)
## Define simulation tables for urine observations
SimTableA0Obs <- tableParams(
name = "SimTableA0Obs.csv",
timesList = c(12, 24),
variablesList = "A0Obs",
forSimulation = TRUE
)
## Simulation setup
SimSetup <- NlmeSimulationParams(
numReplicates = 50,
seed = 1,
simulationTables = c(SimTableCObs, SimTableA0Obs)
)Next, we will run the model using the simulation setup given above, through the simmodel function.
job <- simmodel(model, SimSetup)Simulation Results
The returned job object contains all the simulation
tables defined. We will build plots for both simulated drug
concentration at the central compartment and simulated urine
observations at each dose level specified:
## Simulated drug concentration at the central compartment
dt_CObs <- job$SimTableCObs
setnames(dt_CObs, c("# repl"), c("Replicate"))
dt_CObs$Replicate <- as.factor(dt_CObs$Replicate)
dt_CObs$id5 <- as.factor(dt_CObs$id5)
levels(dt_CObs$id5) = c(paste0("Dose = ", dt_SimInputData$Dose[1]),
paste0("Dose = ", dt_SimInputData$Dose[2]))
## Plot simulated drug concentration at the central compartment
ggplot(dt_CObs, aes(x = time, y = CObs, group = Replicate, color = Replicate)) +
scale_y_log10() +
geom_line() +
geom_point() +
ylab("Drug Concentration \n at the central compartment") +
facet_grid(.~ id5)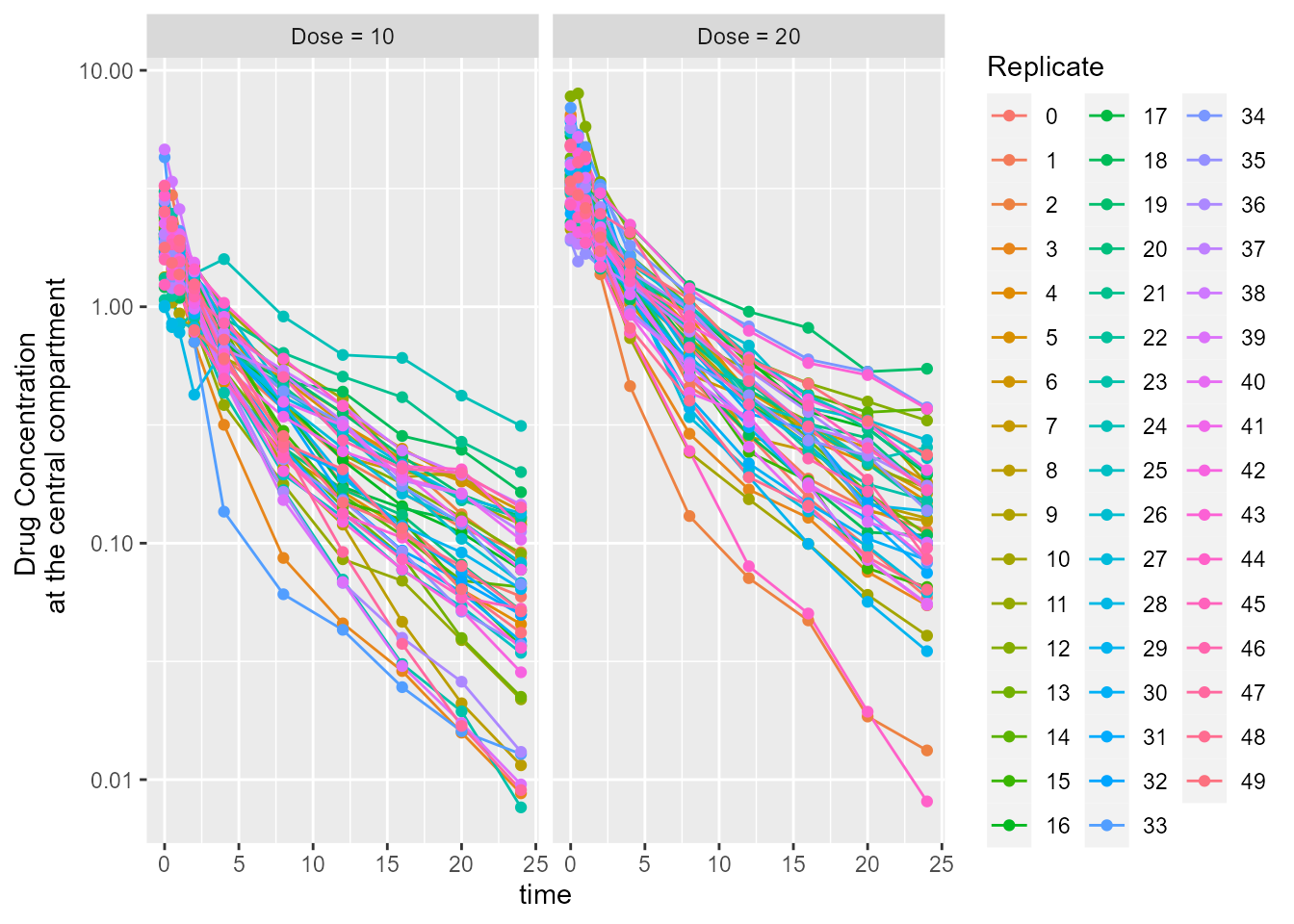
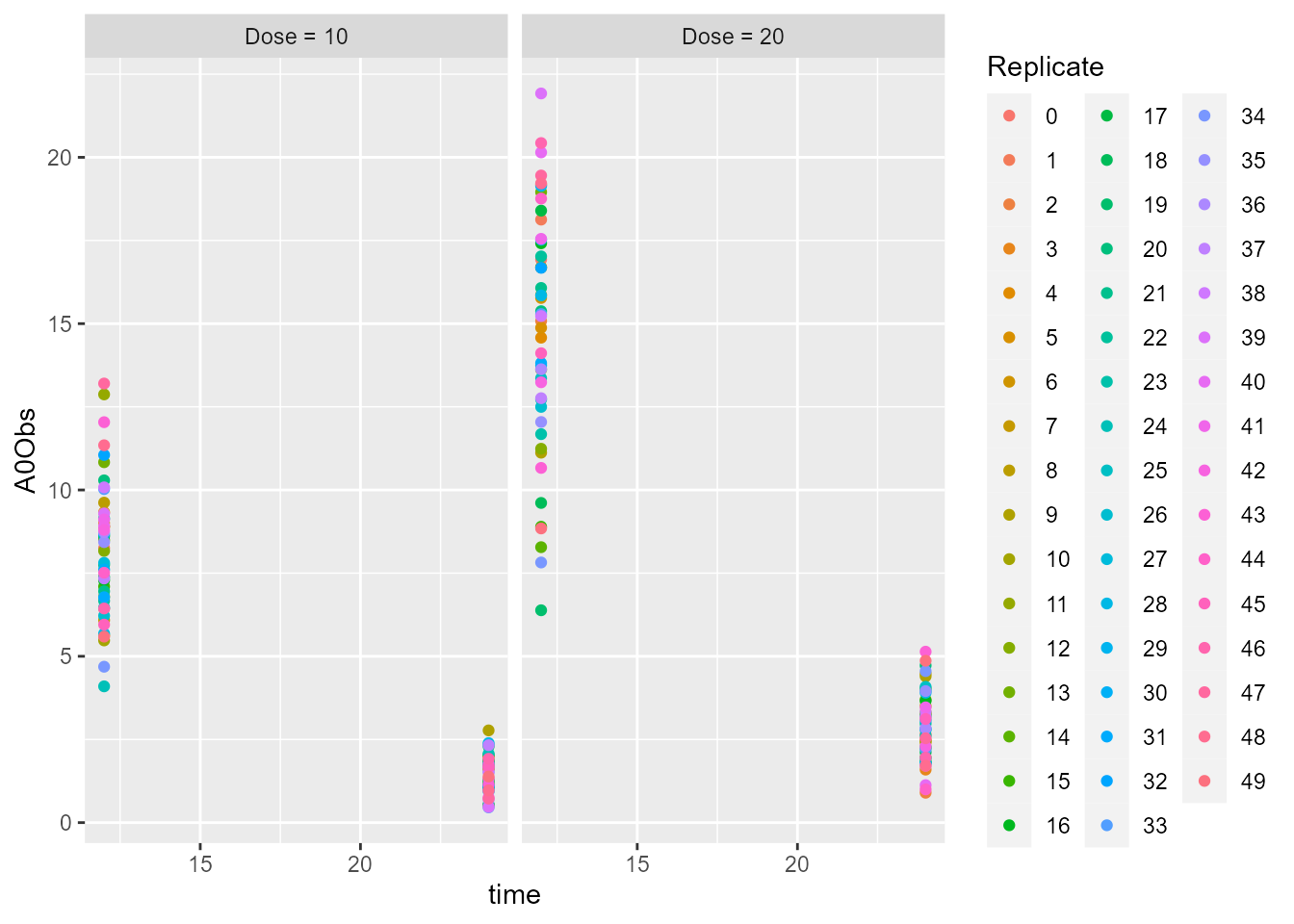
## Simulated urine observations
dt_A0Obs <- job$SimTableA0Obs
setnames(dt_A0Obs, c("# repl"), c("Replicate"))
dt_A0Obs$Replicate <- as.factor(dt_A0Obs$Replicate)
dt_A0Obs$id5 <- as.factor(dt_A0Obs$id5)
levels(dt_A0Obs$id5) = c(paste0("Dose = ", dt_SimInputData$Dose[1]),
paste0("Dose = ", dt_SimInputData$Dose[2]))
## Plot simulated urine observations
ggplot(dt_A0Obs, aes(x = time, y = A0Obs, group = Replicate, color = Replicate)) +
geom_point() +
facet_grid(.~ id5)