Advanced Plot Customization
plot_styling.RmdOverview
Certara.ModelResults uses functions from both the
xpose and Certara.Xpose.NLME packages to
generate model diagnostic plots. The resulting class object returned
from the plotting functions is a ggplot object however,
which means we can use additional functions from the
ggplot2 package to further customize our plot.
Combine multiple plots
Tag GOF plots in resultsUI()
We can combine separate plots that we’ve generated in
resultsUI() into a single plot. To begin, we’ll launch
Certara.ModelResults with our example data
xpdb_NLME and tag the following Goodness of Fit (GOF)
plots: DV vs PRED, DV vs IPRED,
CWRES vs PRED, CWRES vs IVAR.
library(Certara.ModelResults)
resultsUI(xpdb = xpdb_NLME)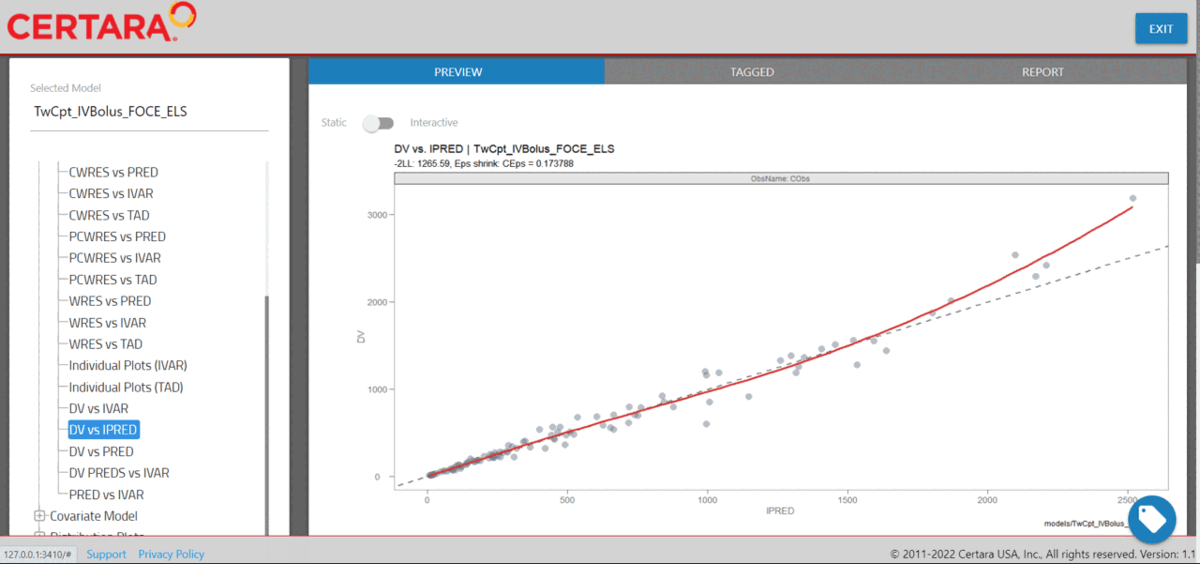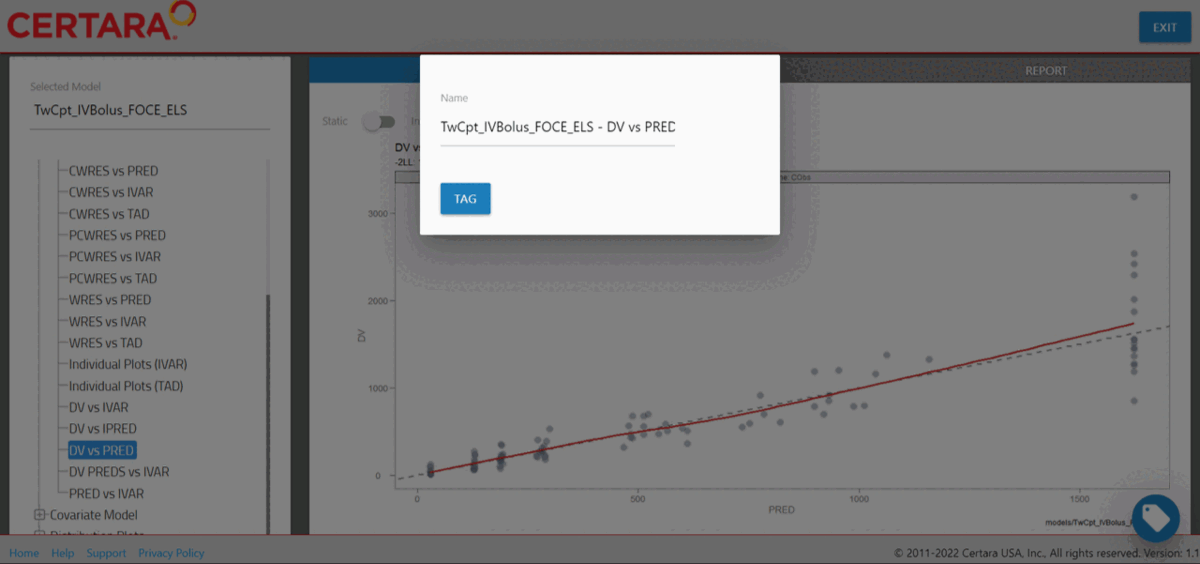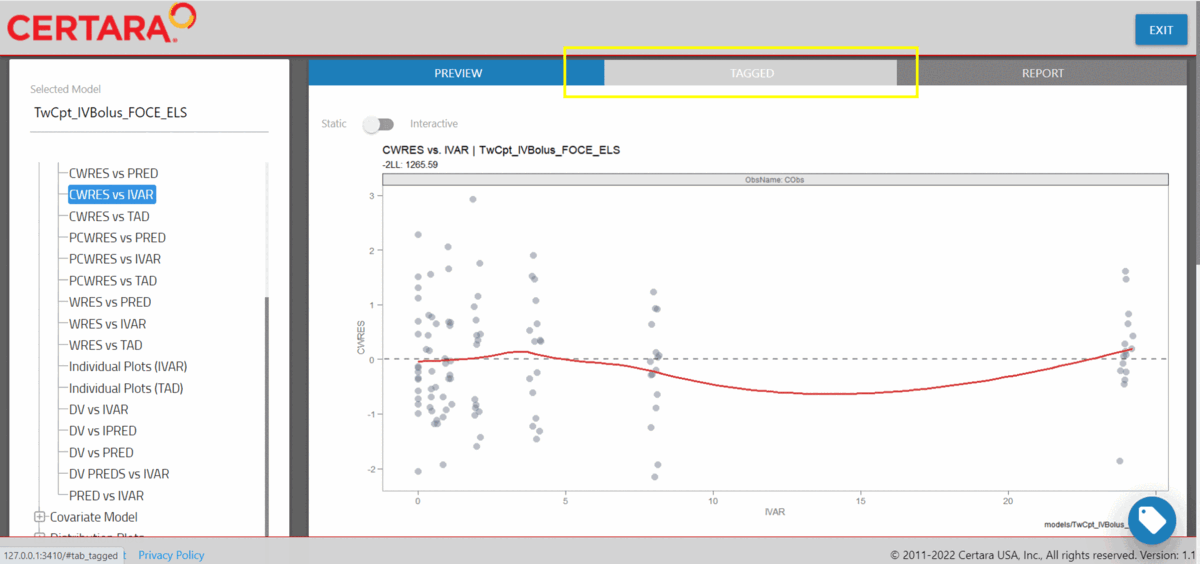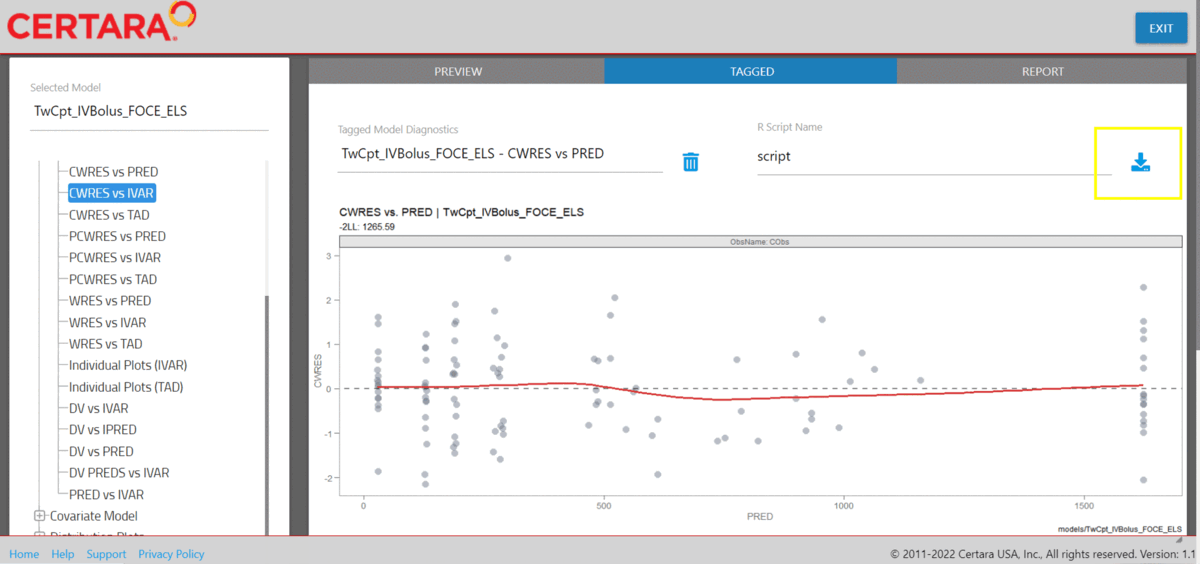
Download R script from Shiny GUI
The following R script GOF_plots.R was downloaded from
the Shiny GUI.
library(Certara.ModelResults)
library(Certara.Xpose.NLME)
library(xpose)
library(ggplot2)
library(dplyr)
library(tidyr)
library(magrittr)
library(flextable)
xpobj <- xpdb_NLME[["TwCpt_IVBolus_SelectedCovariateModel_FOCE_ELS"]]
plot <- xpobj %>%
filter(!is.na(ObsName)) %>%
dv_vs_pred(type = "ps", facets = c("ObsName", NULL), scales = "free", nrow = NULL, ncol = NULL, subtitle = "-2LL: @ofv", page = NULL, guide = TRUE, guide_linetype = "dashed", guide_alpha = 0.5, guide_color = "#000000", guide_linewidth = 1L, smooth_method = "loess", smooth_span = 0.75, smooth_linetype = "solid", smooth_color = "#D63636", smooth_linewidth = 1.2, log = NULL, point_shape = 16, point_color = "#757D8F", point_alpha = 0.5, point_size = 3.9, line_color = "#000000", line_alpha = 0.5, line_linewidth = 1L, line_linetype = "solid") +
theme_certara()
plot
xpobj <- xpdb_NLME[["TwCpt_IVBolus_SelectedCovariateModel_FOCE_ELS"]]
plot <- xpobj %>%
filter(!is.na(ObsName)) %>%
dv_vs_ipred(type = "ps", facets = c("ObsName", NULL), scales = "free", nrow = NULL, ncol = NULL, page = NULL, subtitle = "-2LL: @ofv, Eps shrink: @epsshk", guide = TRUE, guide_linetype = "dashed", guide_alpha = 0.5, guide_color = "#000000", guide_linewidth = 1L, smooth_method = "loess", smooth_span = 0.75, smooth_linetype = "solid", smooth_color = "#D63636", smooth_linewidth = 1.2, log = NULL, point_shape = 16, point_color = "#757D8F", point_alpha = 0.5, point_size = 3.9, line_color = "#000000", line_alpha = 0.5, line_linewidth = 1L, line_linetype = "solid") +
theme_certara()
plot
xpobj <- xpdb_NLME[["TwCpt_IVBolus_SelectedCovariateModel_FOCE_ELS"]]
plot <- xpobj %>%
filter(!is.na(ObsName)) %>%
res_vs_pred(res = "CWRES", type = "ps", facets = c("ObsName", NULL), nrow = NULL, ncol = NULL, scales = "free", subtitle = "-2LL: @ofv", page = NULL, smooth_method = "loess", smooth_span = 0.75, smooth_linetype = "solid", smooth_color = "#D63636", smooth_linewidth = 1.2, log = NULL, guide = TRUE, guide_linetype = "dashed", guide_alpha = 0.5, guide_color = "#000000", guide_linewidth = 1L, point_shape = 16, point_color = "#757D8F", point_alpha = 0.5, point_size = 3.9, line_color = "#000000", line_alpha = 0.5, line_linewidth = 1L, line_linetype = "solid") +
theme_certara()
plot
xpobj <- xpdb_NLME[["TwCpt_IVBolus_SelectedCovariateModel_FOCE_ELS"]]
plot <- xpobj %>%
filter(!is.na(ObsName)) %>%
res_vs_idv(res = "CWRES", type = "ps", facets = c("ObsName", NULL), scales = "free", nrow = NULL, ncol = NULL, subtitle = "-2LL: @ofv", page = NULL, smooth_method = "loess", smooth_span = 0.75, smooth_linetype = "solid", smooth_color = "#D63636", smooth_linewidth = 1.2, log = NULL, guide = TRUE, guide_linetype = "dashed", guide_alpha = 0.5, guide_color = "#000000", guide_linewidth = 1L, point_shape = 16, point_color = "#757D8F", point_alpha = 0.5, point_size = 3.9, line_color = "#000000", line_alpha = 0.5, line_linewidth = 1L, line_linetype = "solid") +
theme_certara()
plotCreate plot objects
To combine the individual plots into a single plot, we’ll assign
these plots new distinct values: plot_dv_pred,
plot_dv_ipred, plot_cwres_pred,
plot_cwres_time.
library(Certara.Xpose.NLME)
library(xpose)
library(ggplot2)
xpobj <- xpdb_NLME[["TwCpt_IVBolus_SelectedCovariateModel_FOCE_ELS"]]
plot_dv_pred <- xpobj %>%
filter(!is.na(ObsName)) %>%
dv_vs_pred(type = "ps", facets = c("ObsName", NULL), scales = "free", nrow = NULL, ncol = NULL, subtitle = "-2LL: @ofv", page = NULL, guide = TRUE, guide_linetype = "dashed", guide_alpha = 0.5, guide_color = "#000000", guide_linewidth = 1L, smooth_method = "loess", smooth_span = 0.75, smooth_linetype = "solid", smooth_color = "#D63636", smooth_linewidth = 1.2, log = NULL, point_shape = 16, point_color = "#757D8F", point_alpha = 0.5, point_size = 3.9, line_color = "#000000", line_alpha = 0.5, line_linewidth = 1L, line_linetype = "solid") +
theme_certara()
plot_dv_ipred <- xpobj %>%
filter(!is.na(ObsName)) %>%
dv_vs_ipred(type = "ps", facets = c("ObsName", NULL), scales = "free", nrow = NULL, ncol = NULL, page = NULL, subtitle = "-2LL: @ofv, Eps shrink: @epsshk", guide = TRUE, guide_linetype = "dashed", guide_alpha = 0.5, guide_color = "#000000", guide_linewidth = 1L, smooth_method = "loess", smooth_span = 0.75, smooth_linetype = "solid", smooth_color = "#D63636", smooth_linewidth = 1.2, log = NULL, point_shape = 16, point_color = "#757D8F", point_alpha = 0.5, point_size = 3.9, line_color = "#000000", line_alpha = 0.5, line_linewidth = 1L, line_linetype = "solid") +
theme_certara()
plot_cwres_pred <- xpobj %>%
filter(!is.na(ObsName)) %>%
res_vs_pred(res = "CWRES", type = "ps", facets = c("ObsName", NULL), nrow = NULL, ncol = NULL, scales = "free", subtitle = "-2LL: @ofv", page = NULL, smooth_method = "loess", smooth_span = 0.75, smooth_linetype = "solid", smooth_color = "#D63636", smooth_linewidth = 1.2, log = NULL, guide = TRUE, guide_linetype = "dashed", guide_alpha = 0.5, guide_color = "#000000", guide_linewidth = 1L, point_shape = 16, point_color = "#757D8F", point_alpha = 0.5, point_size = 3.9, line_color = "#000000", line_alpha = 0.5, line_linewidth = 1L, line_linetype = "solid") +
theme_certara()
plot_cwres_time <- xpobj %>%
filter(!is.na(ObsName)) %>%
res_vs_idv(res = "CWRES", type = "ps", facets = c("ObsName", NULL), scales = "free", nrow = NULL, ncol = NULL, subtitle = "-2LL: @ofv", page = NULL, smooth_method = "loess", smooth_span = 0.75, smooth_linetype = "solid", smooth_color = "#D63636", smooth_linewidth = 1.2, log = NULL, guide = TRUE, guide_linetype = "dashed", guide_alpha = 0.5, guide_color = "#000000", guide_linewidth = 1L, point_shape = 16, point_color = "#757D8F", point_alpha = 0.5, point_size = 3.9, line_color = "#000000", line_alpha = 0.5, line_linewidth = 1L, line_linetype = "solid") +
theme_certara()Combine plots using grid.arrange()
We will use grid.arrange() from the
gridExtra package to arrange our plots into a single
grid.
Note: I have specified fig.width = 8, fig.height = 8
in the Rmd chunk options to reproduce this plot dimension.
library(gridExtra)
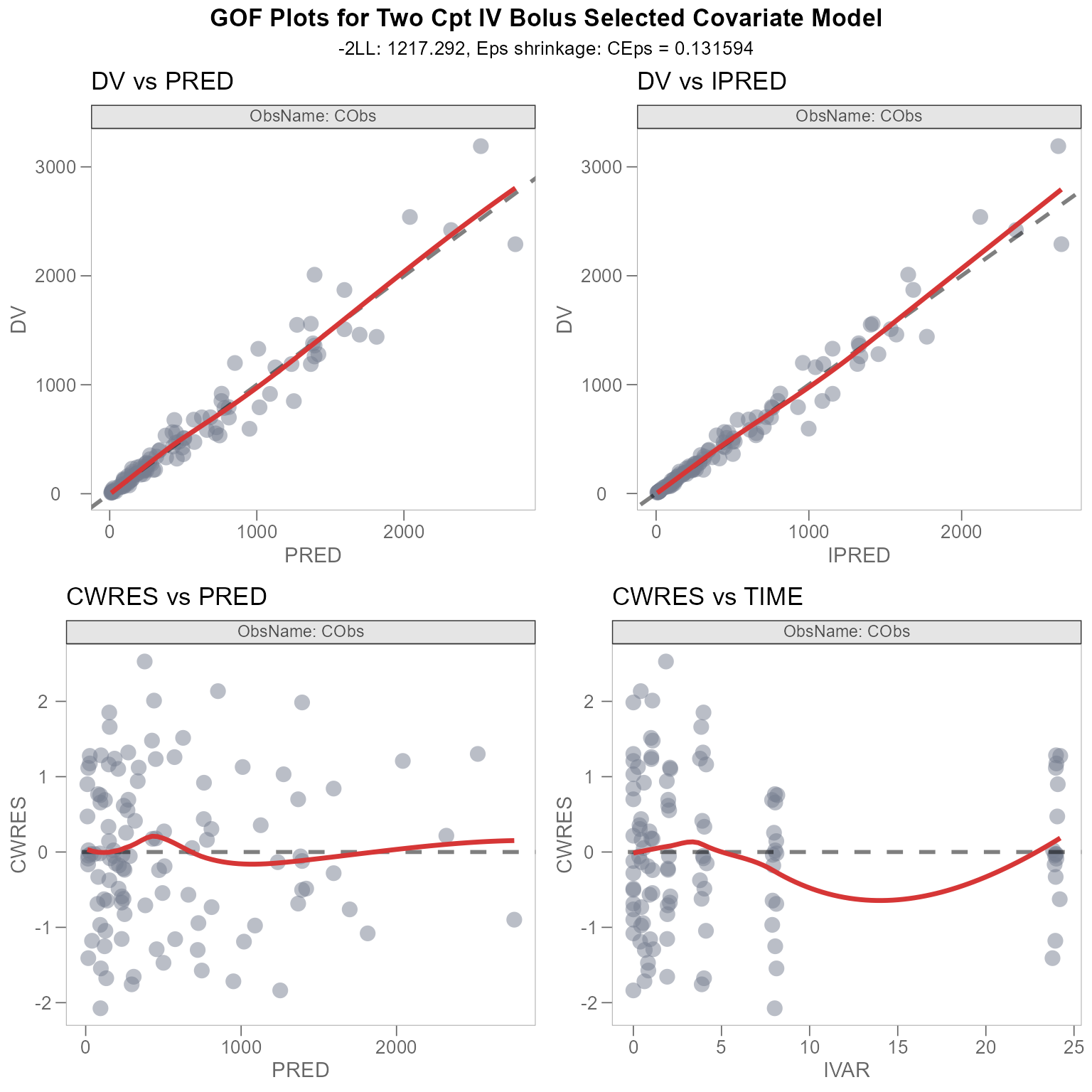
plot_combined_gof <- grid.arrange(plot_dv_pred, plot_dv_ipred, plot_cwres_pred, plot_cwres_time)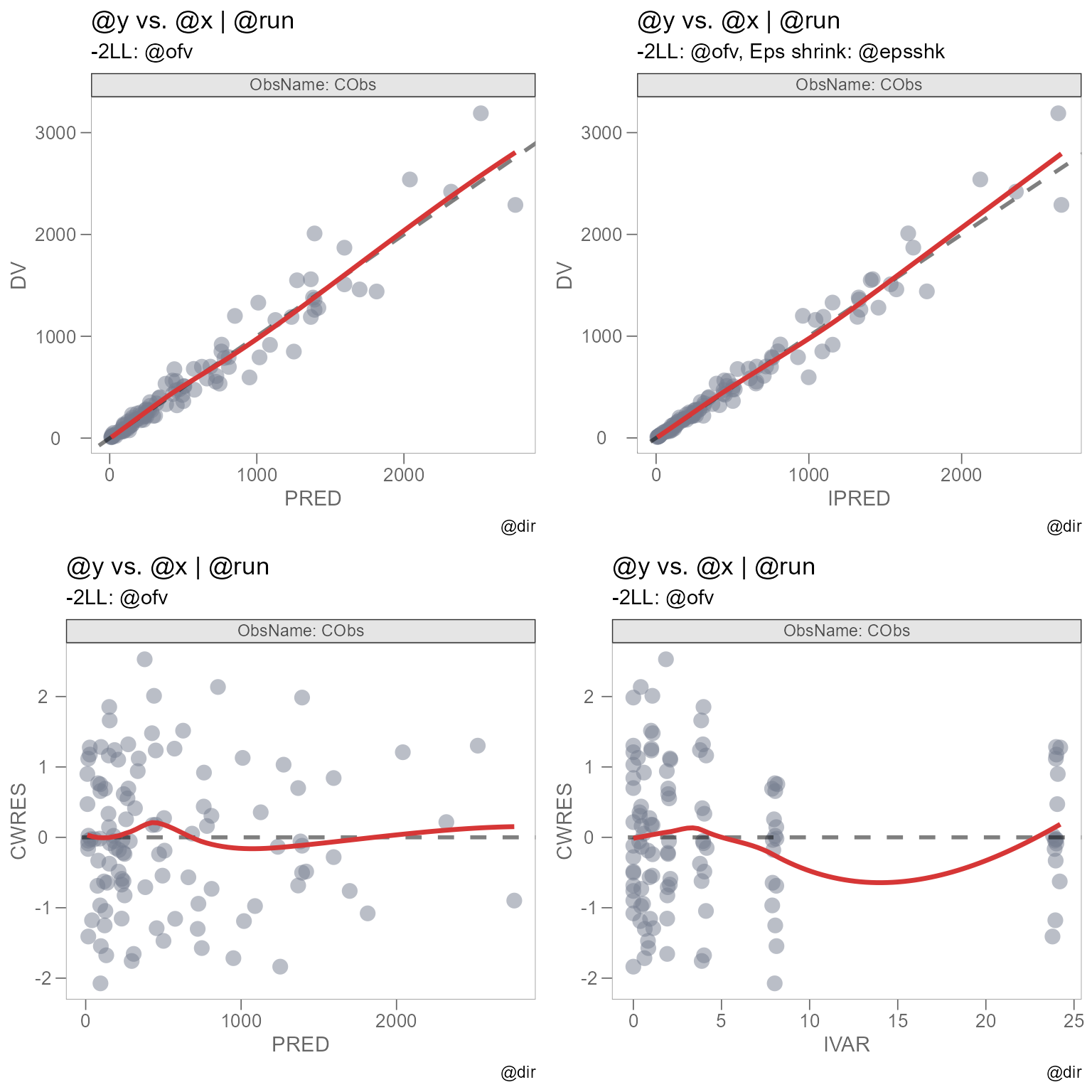
We can see that there are some issues with our plot titles. The
titles and subtitles originally utilized the @ keyword from
the xpose package, which allows us to specify key variables
in our xpose_data object as plot text. See
template_titles().
However, the functionality offered by template_titles()
only works if the plot is of class xpose_plot.
class(plot_dv_pred)
#> [1] "xpose_plot" "gg" "ggplot"We can see that our combined GOF plot is of the following classes:
class(plot_combined_gof)
#> [1] "gtable" "gTree" "grob" "gDesc"To resolve the issue with our titles, we’ll manually change the
titles of our individual plots, without using the @ tag
from template_titles(), since this functionality is not
supported. Let’s also remove the subtitle and caption for each plot.
Note: This can be done in the Shiny GUI. Navigate to the “DISPLAY” subtab under the “PREVIEW” tab and uncheck “Default Text” to change text in the GUI, then tag the plot and generate the corresponding R code.
plot_dv_pred <- plot_dv_pred +
labs(title = "DV vs PRED", subtitle = NULL, caption = NULL)
plot_dv_ipred <- plot_dv_ipred +
labs(title = "DV vs IPRED", subtitle = NULL, caption = NULL)
plot_cwres_pred <- plot_cwres_pred +
labs(title = "CWRES vs PRED", subtitle = NULL, caption = NULL)
plot_cwres_time <- plot_cwres_time +
labs(title = "CWRES vs TIME", subtitle = NULL, caption = NULL)Now, we’ll arrange our combined GOF plots once again.
plot_combined_gof <- grid.arrange(plot_dv_pred, plot_dv_ipred, plot_cwres_pred, plot_cwres_time)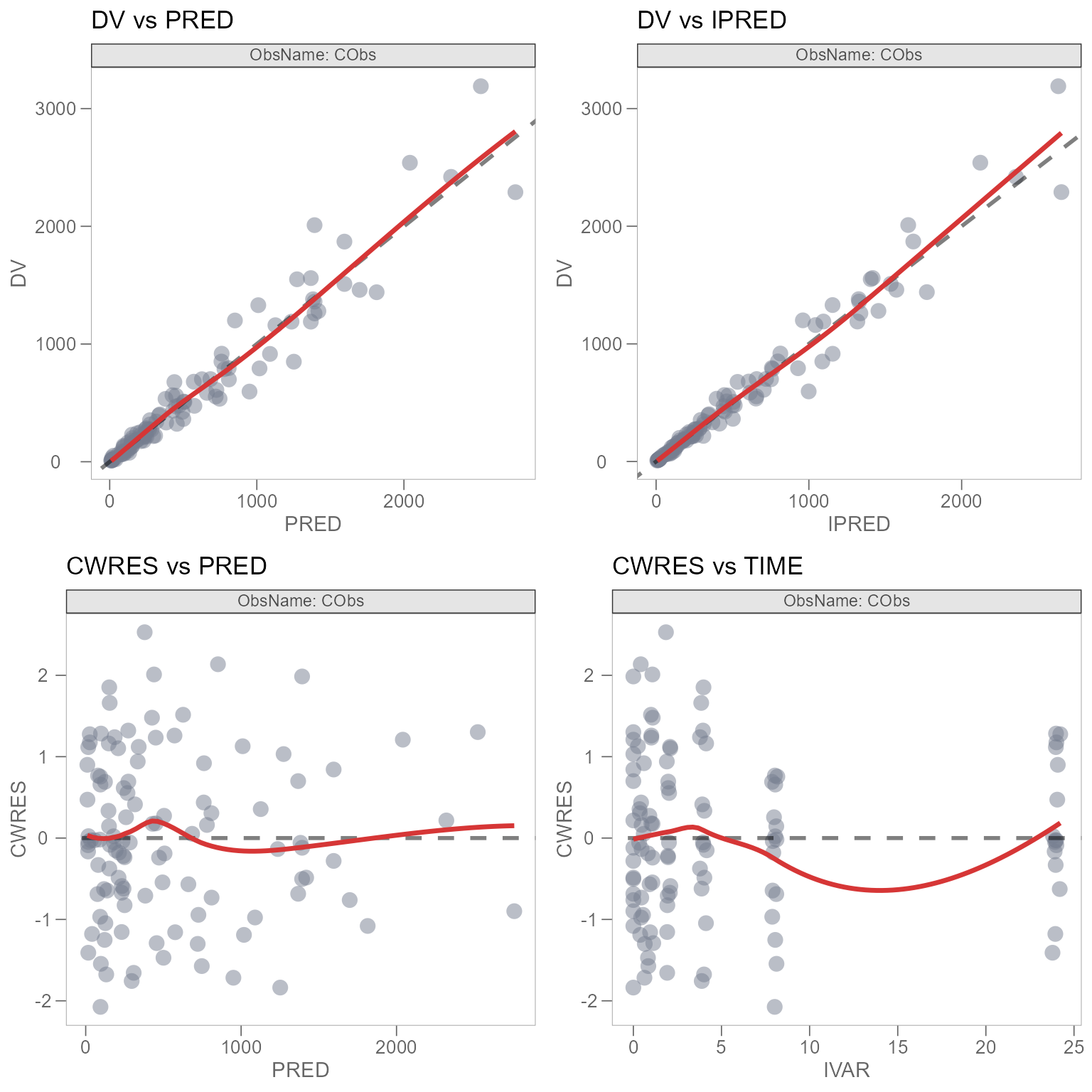
Our plot_combined_gof is looking much better now. But
what if we want to overlay an additional title and subtitle on the
combined GOF plot? We can do so by combining a rectGrob and
textGrob() from the grid package.
Using textGrob() can be sufficient on its own in some
use cases, but this function is unable to add a background color to the
text. This means that a report rendered on a dark mode HTML
page for example may be hard to read with the text and background being
similar colors. To compensate for this, we can create a container for
the text using rectGrob. This will create a rectangular
division that we can set to the same width and color as our plot. Within
this container, we’ll define a textGrob() for both the
title and subtitle. We will extract the ofv and
epsshk values contained in our xpobj and
define them in the subtitle.
library(grid)
library(gridExtra)
library(dplyr)
ofv <- xpobj$summary %>%
filter(problem == 1 & label == "ofv") %>%
select(value)
eps_shk <- xpobj$summary %>%
filter(problem == 1 & label == "epsshk") %>%
select(value)
# Title
title_grob <- grobTree(
rectGrob(width = unit(1, "npc"), gp=gpar(fill = "white", col = NA)),
textGrob('GOF Plots for Two Cpt IV Bolus Selected Covariate Model', gp = gpar(fontsize = 13, fontface = 'bold'))
)
# Subtitle
subtitle_grob <- grobTree(
rectGrob(width = unit(1, "npc"), gp=gpar(fill = "white", col = NA)),
textGrob(paste0("-2LL: ", ofv, ", Eps shrinkage: ", eps_shk), gp = gpar(fontsize = 10))
)
Finally, we’ll use the grid.arrange() function once
more, except now we will arrange our title_grob and
subtitle_grob with our plot_combined_gof. We
will also specify a margin in order to correctly format the
spacing for our title_grob and substitle_grob
grob() objects, using the heights
argument.
# Define margin
margin <- unit(0.5, "line")
# Update combined GOF plot with title and subtitle
updated_plot_combined_gof <- grid.arrange(title_grob,
subtitle_grob,
plot_combined_gof,
heights = unit.c(grobHeight(title_grob) + 2.5*margin,
grobHeight(subtitle_grob) + 2.5*margin,
unit(1,"null")))