Using Model Executor from RStudio
model-executor-rsnlme.RmdOverview
The Certara.RsNLME.ModelExecutor Shiny GUI can be used
alongside Certara.RsNLME for a robust and efficient
execution of RsNLME models. Specifically, users create models and hosts
in R through the Certara.RsNLME package, and execute them
using Certara.RsNLME.ModelExecutor, which allows users to
choose different run modes and specify engine arguments in the Shiny
GUI. All model execution results can be found in the working directory
of the model.
Data
We will be using the built-in pkData from the
Certara.RsNLME package.
library(Certara.RsNLME)
pkData <- Certara.RsNLME::pkData
head(pkData)
#> # A tibble: 6 × 8
#> Subject Nom_Time Act_Time Amount Conc Age BodyWeight Gender
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr>
#> 1 1 0 0 25000 2010 22 73 male
#> 2 1 1 0.26 NA 1330 22 73 male
#> 3 1 1 1.1 NA 565 22 73 male
#> 4 1 2 2.1 NA 216 22 73 male
#> 5 1 4 4.13 NA 180 22 73 male
#> 6 1 8 8.17 NA 120 22 73 maleBuild base model
We will define a basic two-compartment population PK model with IV
bolus as well as its associated column mappings through the
Certara.RsNLME built-in function pkmodel().
The input data used in this example is the built-in pkData
from the Certara.RsNLME package.
*Note: The corresponding R script used in this example can be found in RsNLME Example12
library(Certara.RsNLME.ModelBuilder)
baseModel <- pkmodel(numCompartments = 2, data = pkData,
ID = "Subject", Time = "Act_Time", A1 = "Amount", CObs = "Conc",
modelName = "TwCpt_IVBolus_FOCE_ELS")Next, we will update parameters in our model:
- Disable the corresponding random effects for structural parameter V2
- Change initial values for fixed effects, tvV, tvCl, tvV2, and tvCl2, to be 15, 5, 40, and 15, respectively
- Change the covariance matrix of random effects, nV, nCl, and nCl2, to be a diagonal matrix with all its diagonal elements being 0.1
- Change the standard deviation of residual error to be 0.2
baseModel <- baseModel %>%
structuralParameter(paramName = "V2", hasRandomEffect = FALSE) %>%
fixedEffect(effect = c("tvV", "tvCl", "tvV2", "tvCl2"), value = c(15, 5, 40, 15)) %>%
randomEffect(effect = c("nV", "nCl", "nCl2"), value = rep(0.1, 3)) %>%
residualError(predName = "C", SD = 0.2)Define hosts
We will create two NlmeParallelHost: one for local
execution and the other for remote execution.
host_local <- hostParams(sharedDirectory = Sys.getenv("NLME_ROOT_DIRECTORY"),
parallelMethod = "LOCAL_MPI",
hostName = "local",
numCores = 4)
host_remote <- hostParams(sharedDirectory = "/home/nlmeuser",
parallelMethod = "TORQUE",
hostName = "remote_torque",
machineName = "192.168.15.21",
userName = "nlmeuser",
userPassword = "password",
hostType = "UBUNTU",
isLocal = FALSE,
numCores = 4)
hosts <- c(host_local, host_remote)Fit base model through model executor
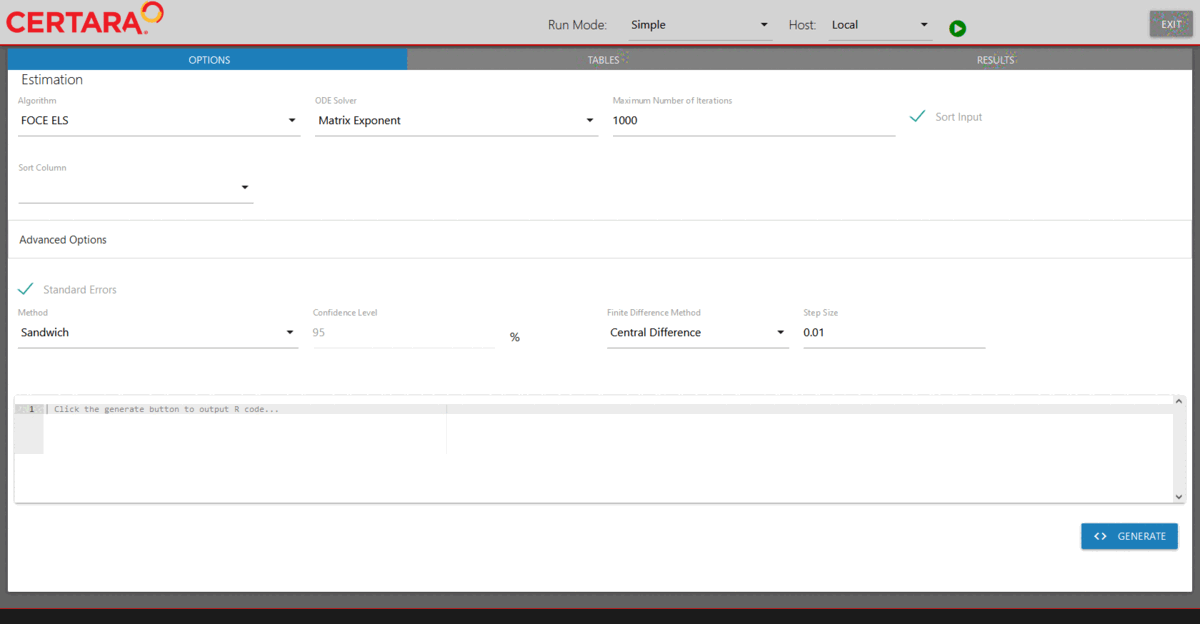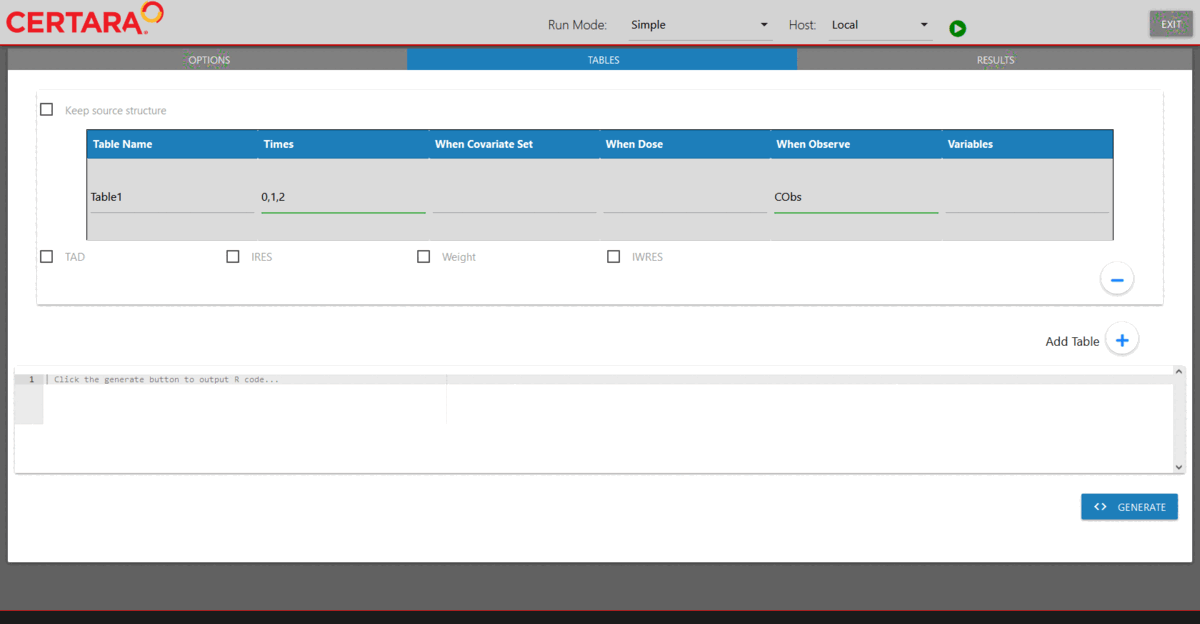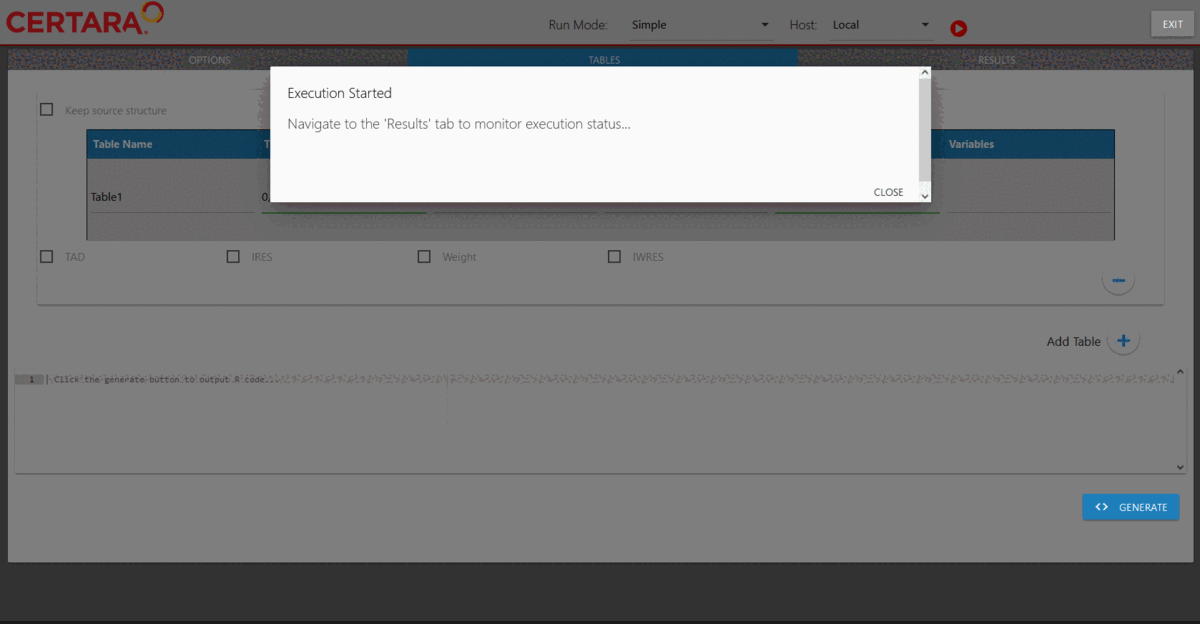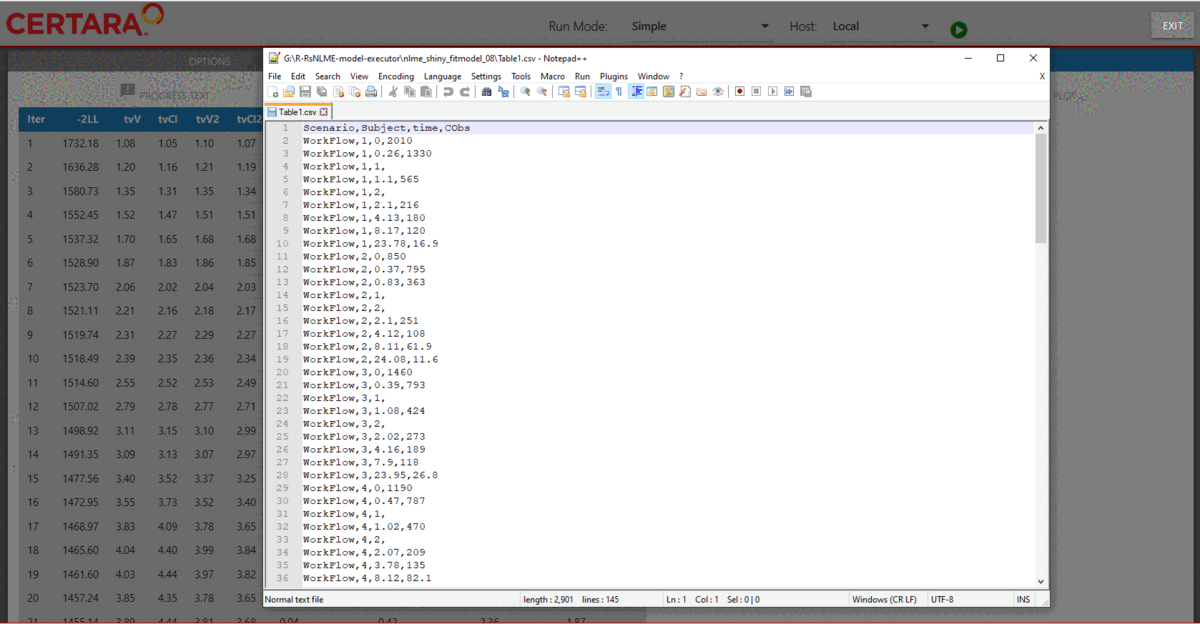
Run model executor using the previously created model and hosts, and execute the model with default setups.
Note: The default values for the NLME engine arguments are chosen
based on the default values of engineParams. Type
?Certara.RsNLME::engineParams for details.
modelExecutorUI(baseModel, hosts)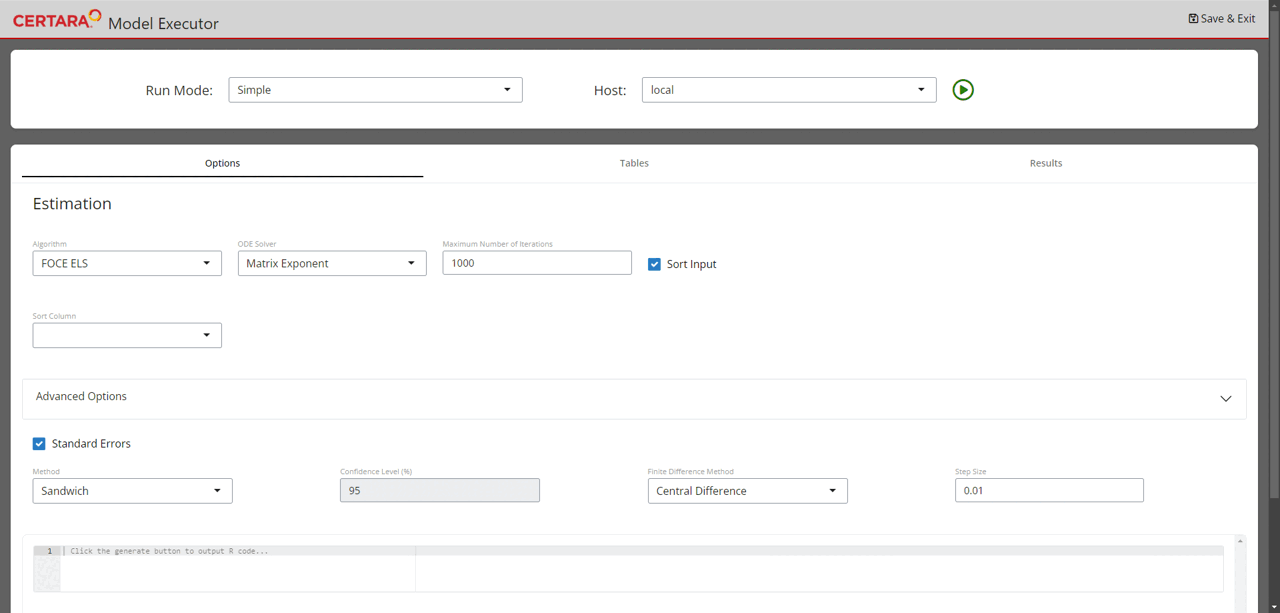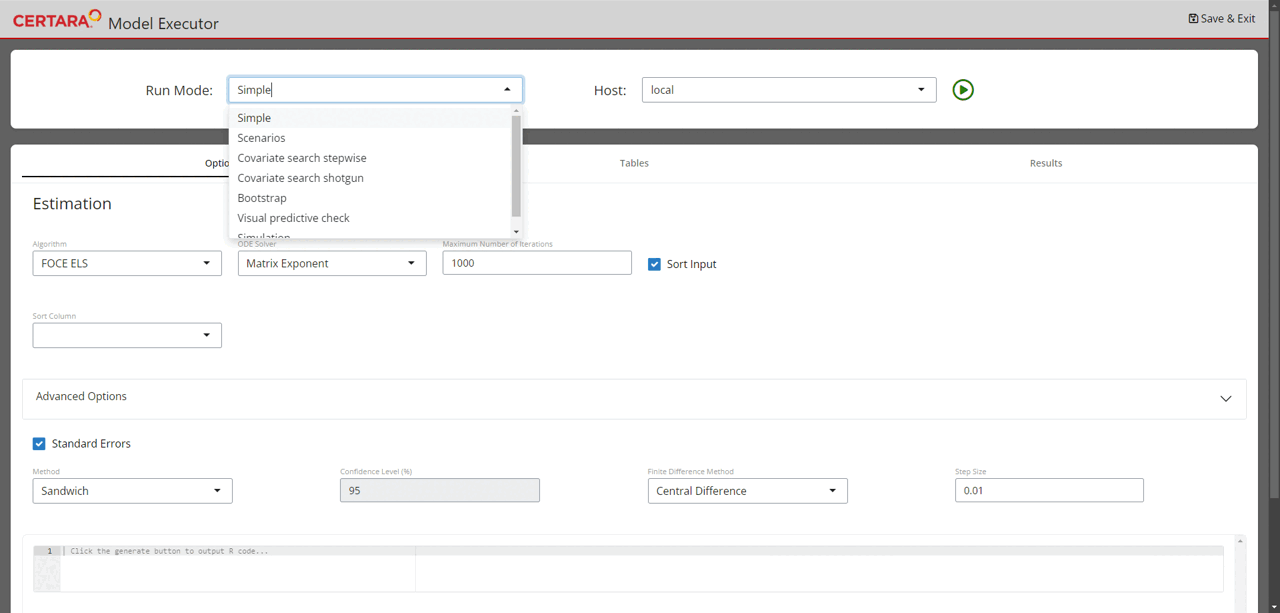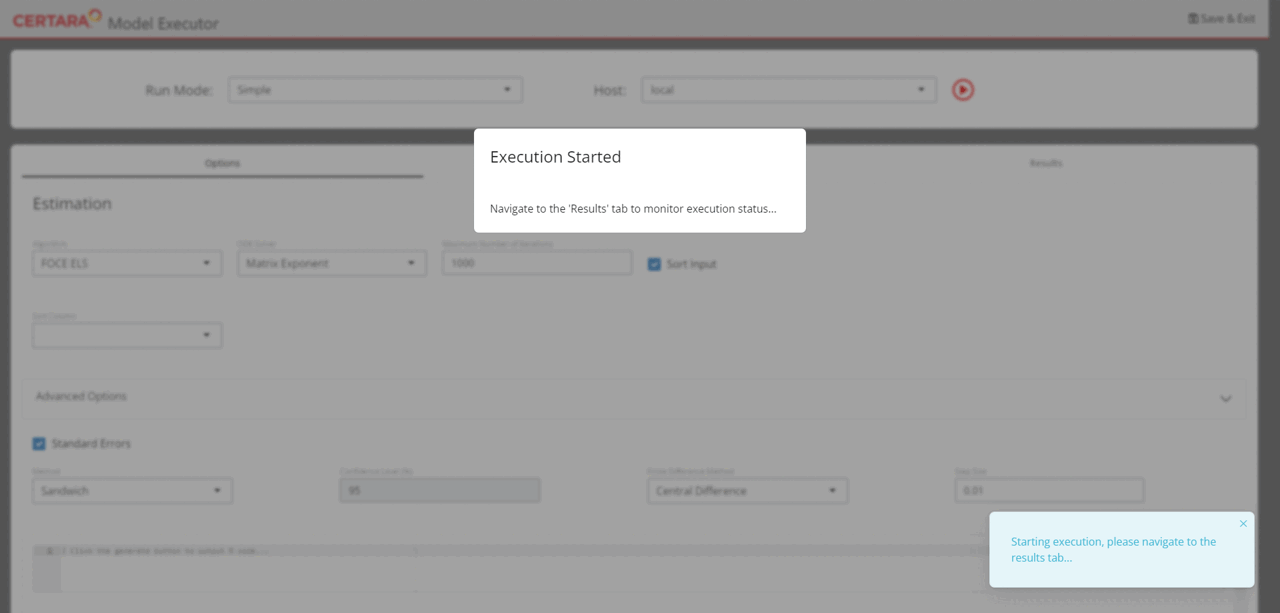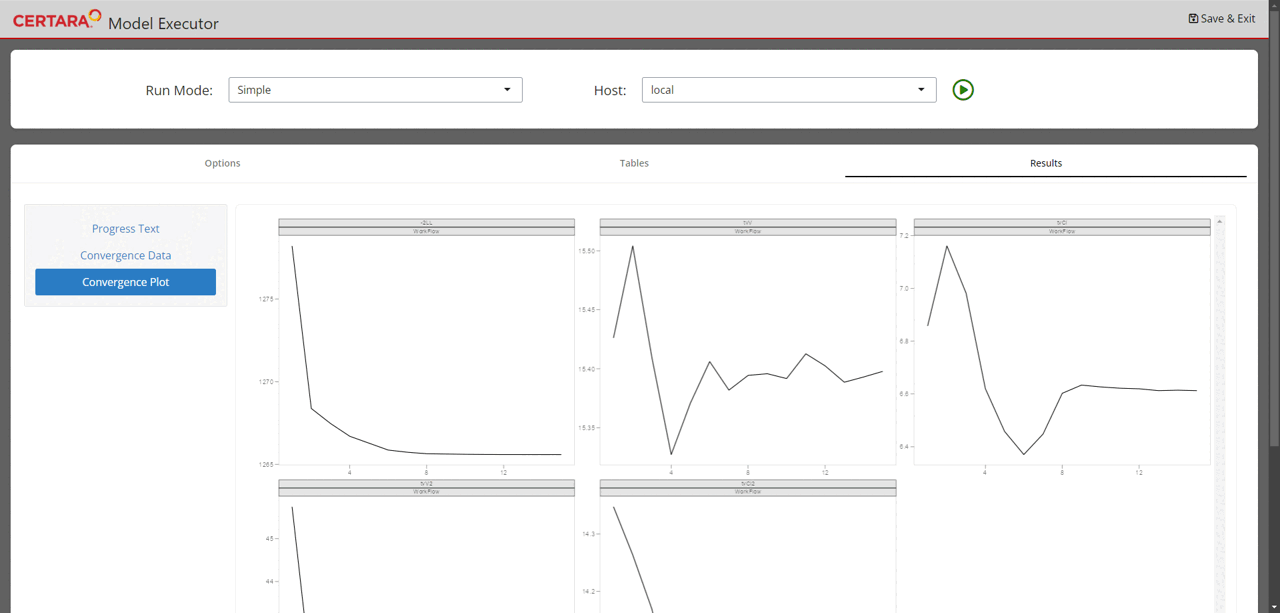
Since we did not provide a value for the wd argument,
results will be written to the current session’s working directory. For
each execution, Model Executor creates a new directory with its name
having the format
nlme_shiny_typeOfRun_numOfSameTypeRunExecution. Since this
is the first time that we are running Model Executor to fit a model,
results for this run can be found under the
nlme_shiny_fitmodel_01 folder.
Real-time convergence plots
For the Simple run mode without any columns selected as Sort, real-time convergence plots are being created. To observe them, navigate to the Results tab (as shown on the above GIF) and switch to Convergence Plot subtab. Plots are being updated during execution and can be used to understand whether the process is converging and if there is a need to adjust execution setup.
Add additional tables to be outputted
For the Simple run mode demonstrated above, one can click the “Add Table” button (located in the lower right corner of the page) to add any desired table to be outputted.
Build covariate model
After fitting the baseModel, let us begin to introduce covariates. We
will first copy our baseModel using the copyModel()
function with acceptAllEffects set to TRUE to accept
parameter estimates from our simple estimation run.
## Set the working directory of baseModel to the one returned by the model executor
baseModel@modelInfo@workingDir <- paste0(getwd(), "/nlme_shiny_fitmodel_01")
## Copy the baseModel with acceptAllEffects = TRUE to accept parameter estimates from its simple estimation run
covariateModel <- copyModel(baseModel, acceptAllEffects = TRUE,
modelName = "TwCpt_IVBolus_CovariateSearch")Next, we will add the continuous covariate BodyWeight to
our model and set the covariate effect on structural parameters
V and Cl.
covariateModel <- addCovariate(covariateModel,
covariate = "BodyWeight",
type = "Continuous",
direction = "Backward",
center = "Mean",
effect = c("V", "Cl"))Run stepwise covariate search for covariate model through model executor
Launch model executor again using the covariate model and hosts created above, choose “Covariate search stepwise” from the drop-down list of Run Mode (located in the top left corner of the page), and then run it.
modelExecutorUI(covariateModel, hosts)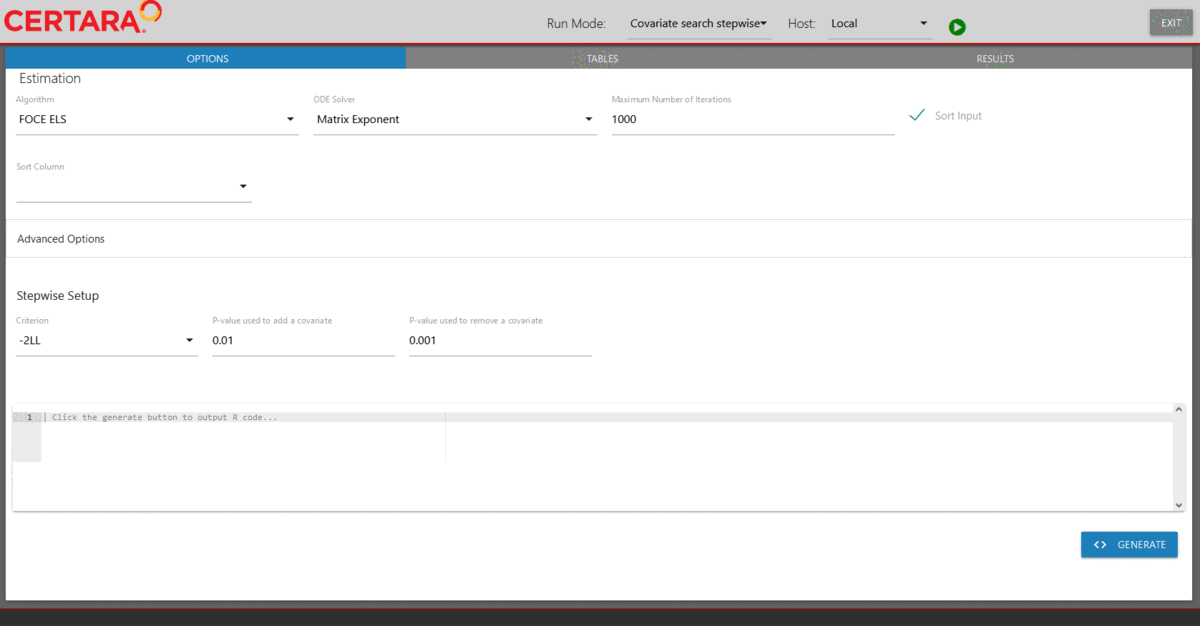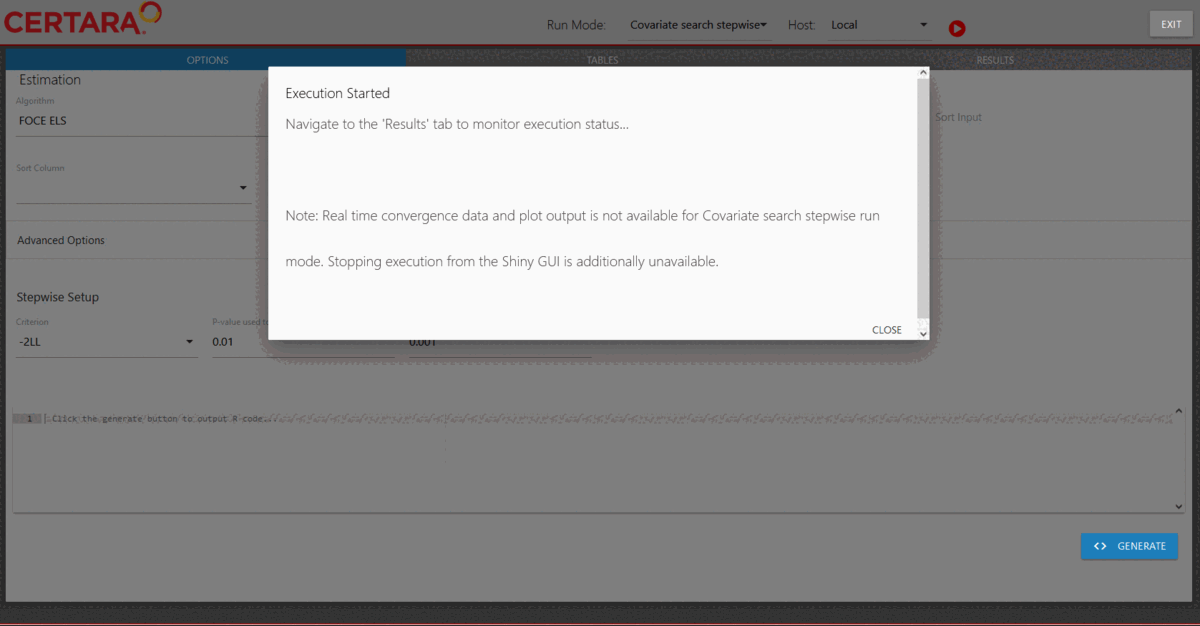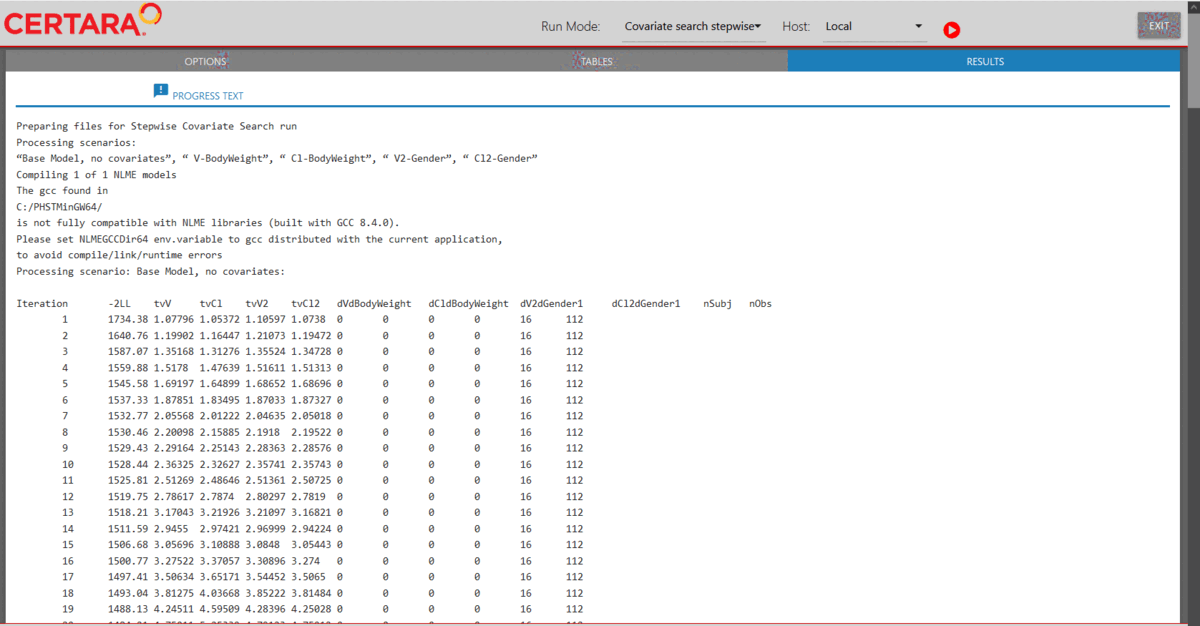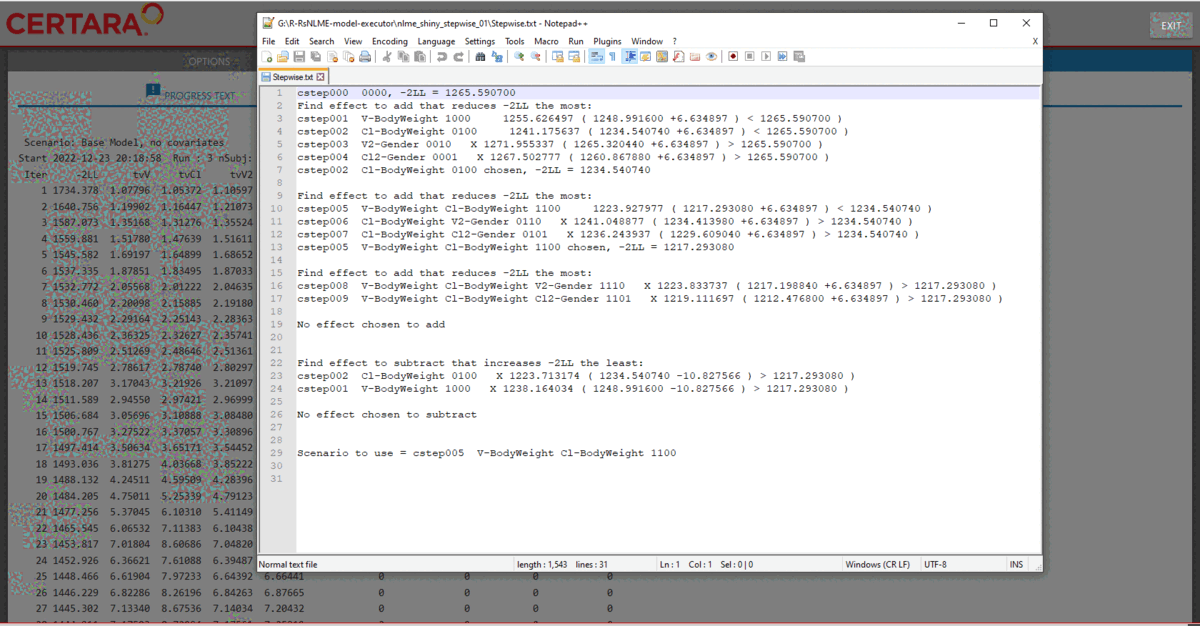
The stepwise.txt file (which can be found in the
nlme_shiny_stepwise_01 directory inside your session’s
working directory) shows that the model selected by stepwise covariate
search is the one with covariate BodyWeight having effect
on both V and Cl.
Fit the selected model through model executor
We will fit the selected model through model executor. First, we will
copy our covariateModel using the copyModel()
function.
selectedCovariateModel <- copyModel(covariateModel,
modelName = "TwCpt_IVBolus_selectedCovariateModel")Next, we will launch model executor using the
selectedCovariateModel and hosts created previously.
modelExecutorUI(selectedCovariateModel, hosts)Then, we will run it using the default setups. Note that this is the
second time that the model executor is used to fit a model. Thus, the
estimation results obtained for this run can be found in the
nlme_shiny_fitmodel_02 folder.
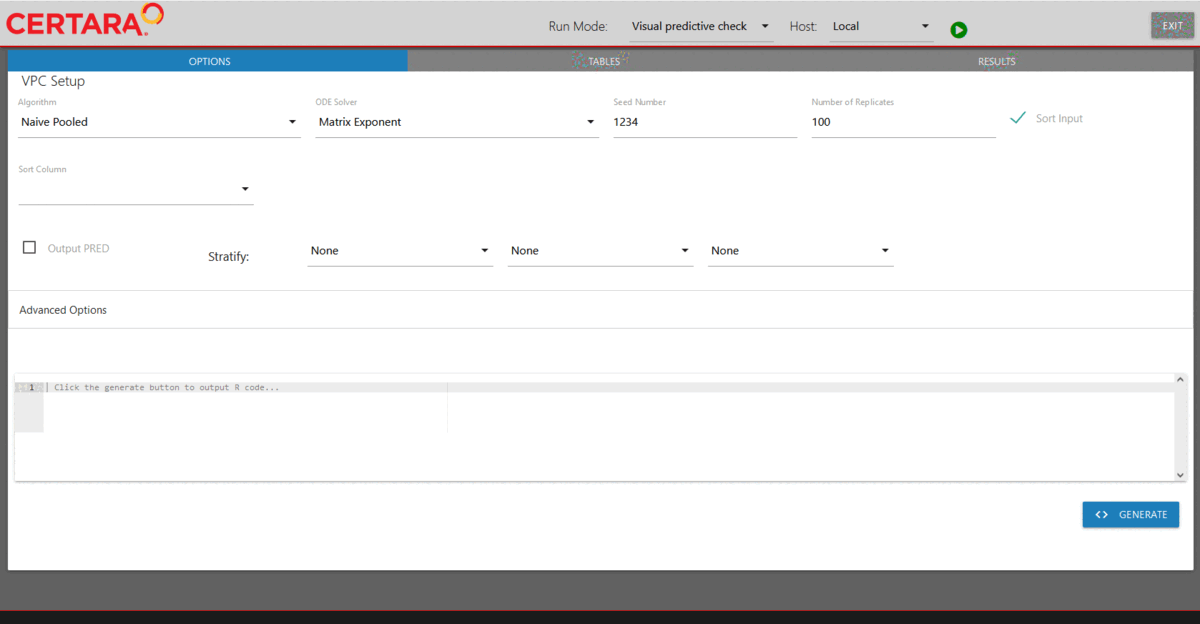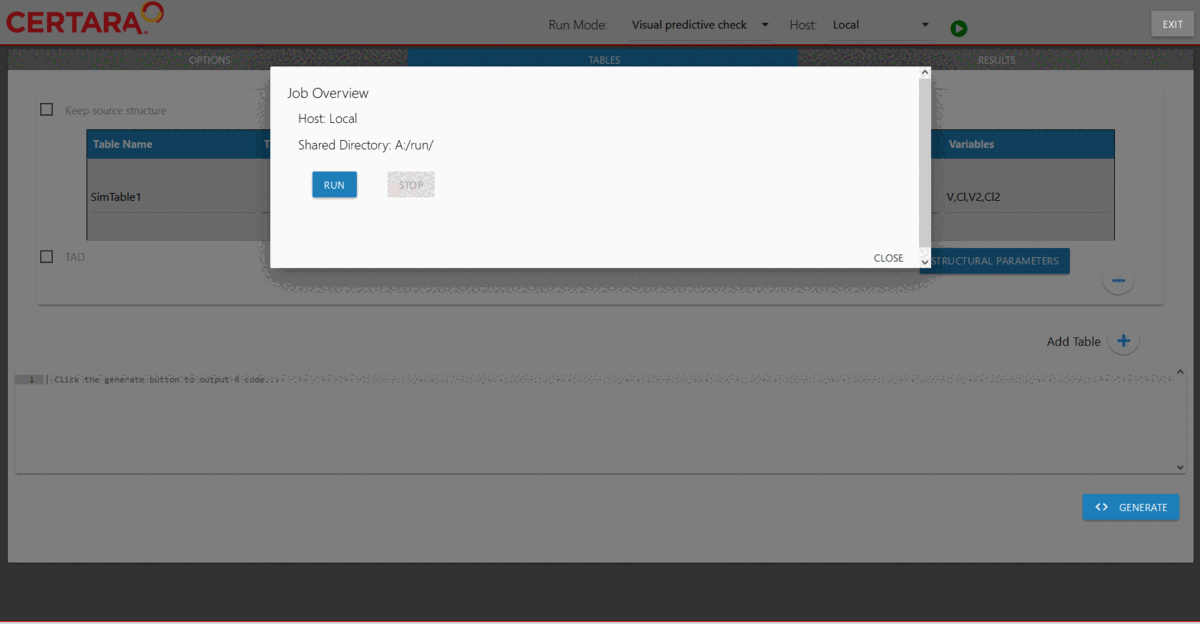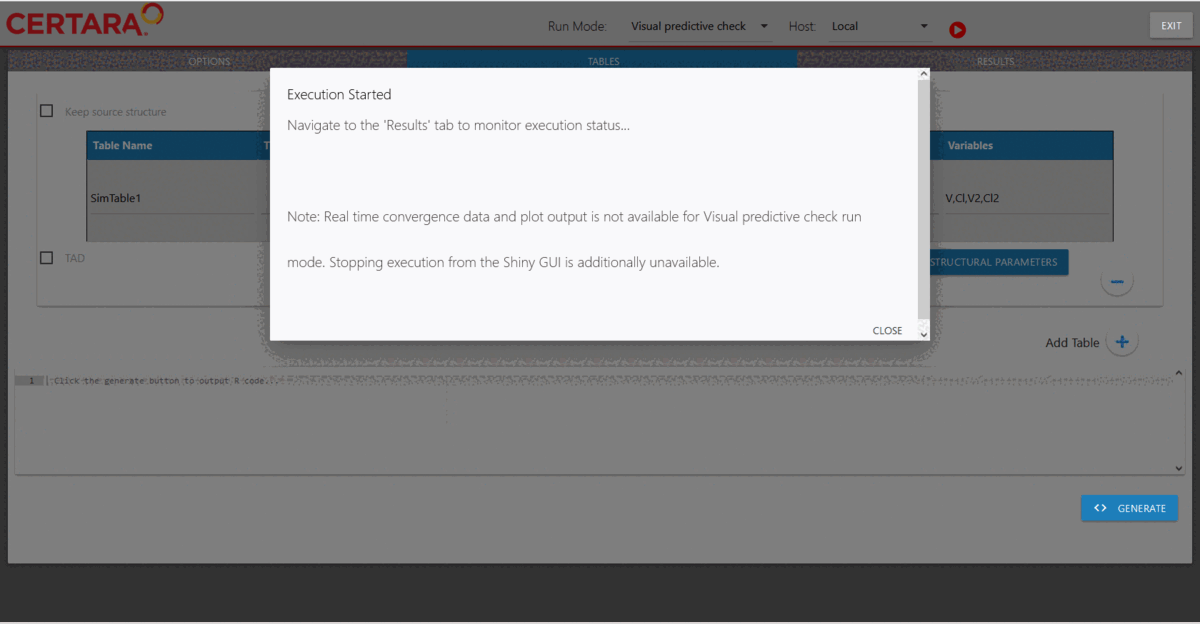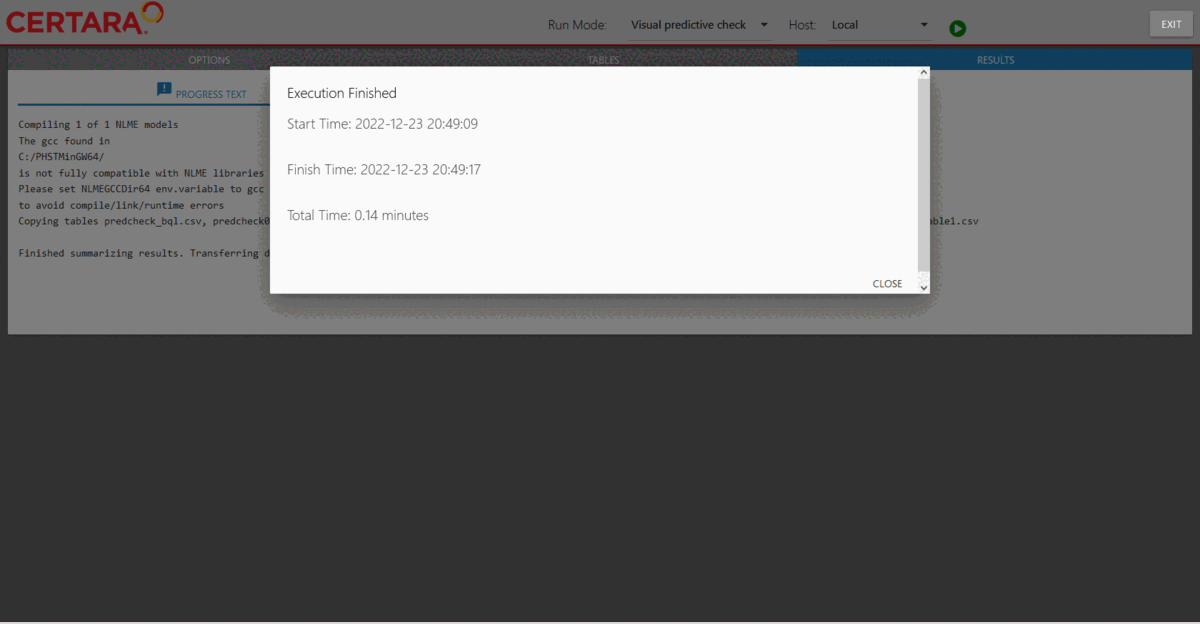
Run VPC for the selected model through model executor
After fitting the selectedCovariateModel, we will run
VPC for it. Again, we will first copy our
selectedCovariateModel using the copyModel()
function with acceptAllEffects set to TRUE to accept
parameter estimates from our simple estimation run.
## Set the working directory of selectedCovariateModel to the one returned by the model executor
selectedCovariateModel@modelInfo@workingDir <- paste0(getwd(), "/nlme_shiny_fitmodel_02")
## Copy the selectedCovariateModel with acceptAllEffects = TRUE to accept parameter estimates from its simple estimation run
vpcSelectedCovariateModel <- copyModel(selectedCovariateModel,
acceptAllEffects = TRUE,
modelName = "TwCpt_IVBolus_slectedCovariateModel_VPC")Next, we will launch model executor using the above model and the
hosts created previously. Then, we will choose “Visual predictive check”
from the drop-down list of Run Mode (located in the top left corner of
the page), add additional simulation tables if desired, and run it.
Results of the VPC run can be found under nlme_shiny_vpc_01
inside your session’s working directory.