tidyvpc 1.5.0
Plot Percentage of BLQ and/or ALQ
For VPC with censoring, users can supply additional arguments to
plot.tidyvpcobj e.g., censoring.type (options
are 'none', 'blq', 'alq', or
'both', defaults to 'none') and
censoring.output (options are 'grid' or
'list', defaults to 'grid').
If censoring.output = 'grid', the plots will be arranged
into single grid plot. Users may pass additional arguments via ellipsis
to egg::ggarrange e.g., nrow = 1,
ncol = 2 in order to customize plots in grid
arrangement.
If censoring.output = 'list', the resulting plots will
be returned individually as elements in list.
Example usage is below:
library(tidyvpc)
#> tidyvpc is part of Certara.R!
#> Follow the link below to learn more about PMx R package development at Certara.
#> https://certara.github.io/R-Certara/
obs_data <- obs_data[MDV == 0]
sim_data <- sim_data[MDV == 0]
obs_data$LLOQ <- obs_data[, ifelse(STUDY == "Study A", 50, 25)]
obs_data$ULOQ <- obs_data[, ifelse(STUDY == "Study A", 125, 100)]
vpc <- observed(obs_data, x = TIME, y = DV) |>
simulated(sim_data, y = DV) |>
censoring(blq = DV < LLOQ, lloq = LLOQ, alq = DV > ULOQ, uloq = ULOQ) |>
stratify(~ STUDY) |>
binning(bin = NTIME) |>
vpcstats(qpred = c(0.1, 0.5, 0.9))If blq data, users may supply
censoring.type = "blq":
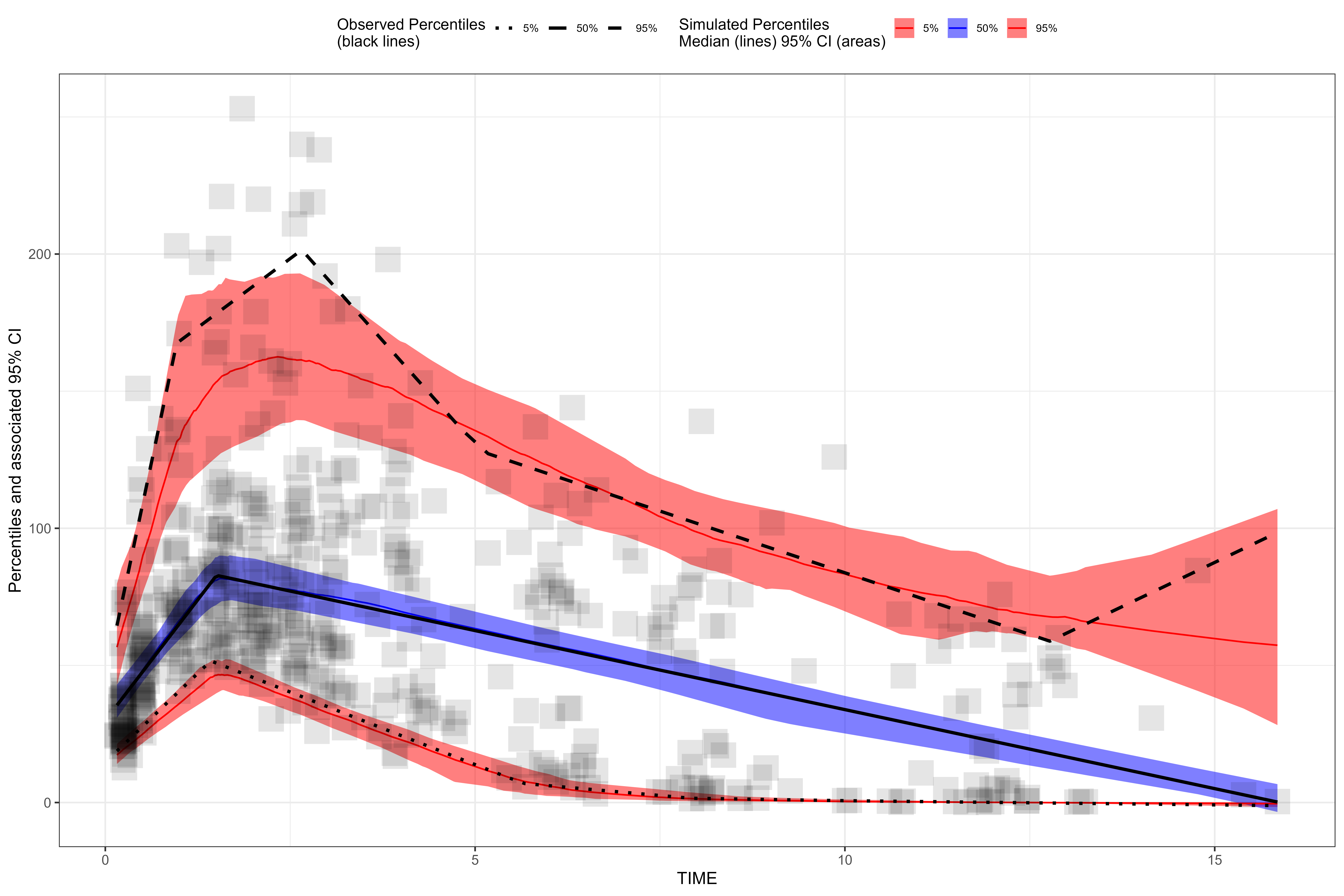
plot(vpc, censoring.type = "blq", censoring.output = "grid", facet.scales = "fixed")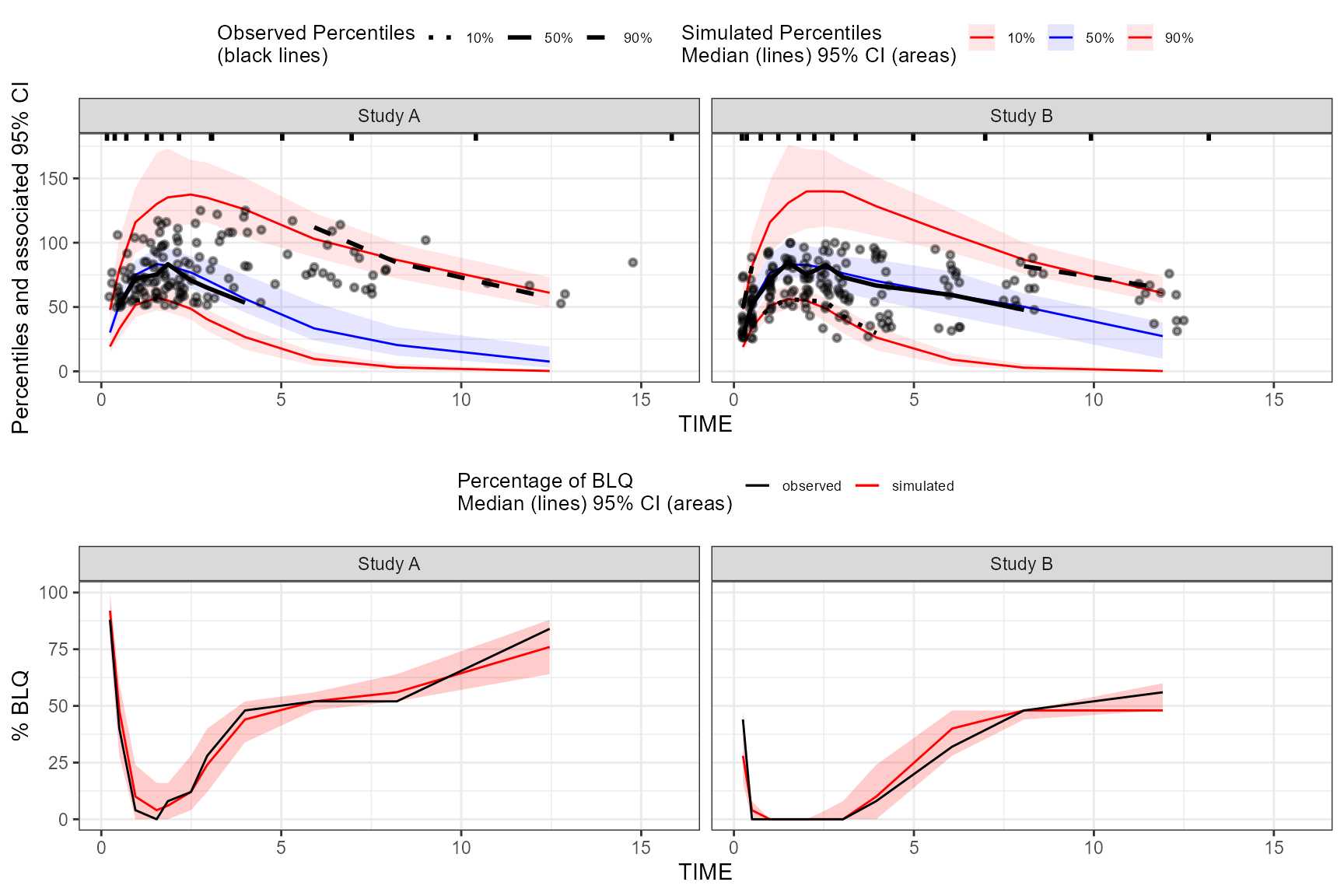
If alq data, users may supply
censoring.type = "alq":
plot(vpc, censoring.type = "alq", censoring.output = "grid", ncol = 2, nrow = 1)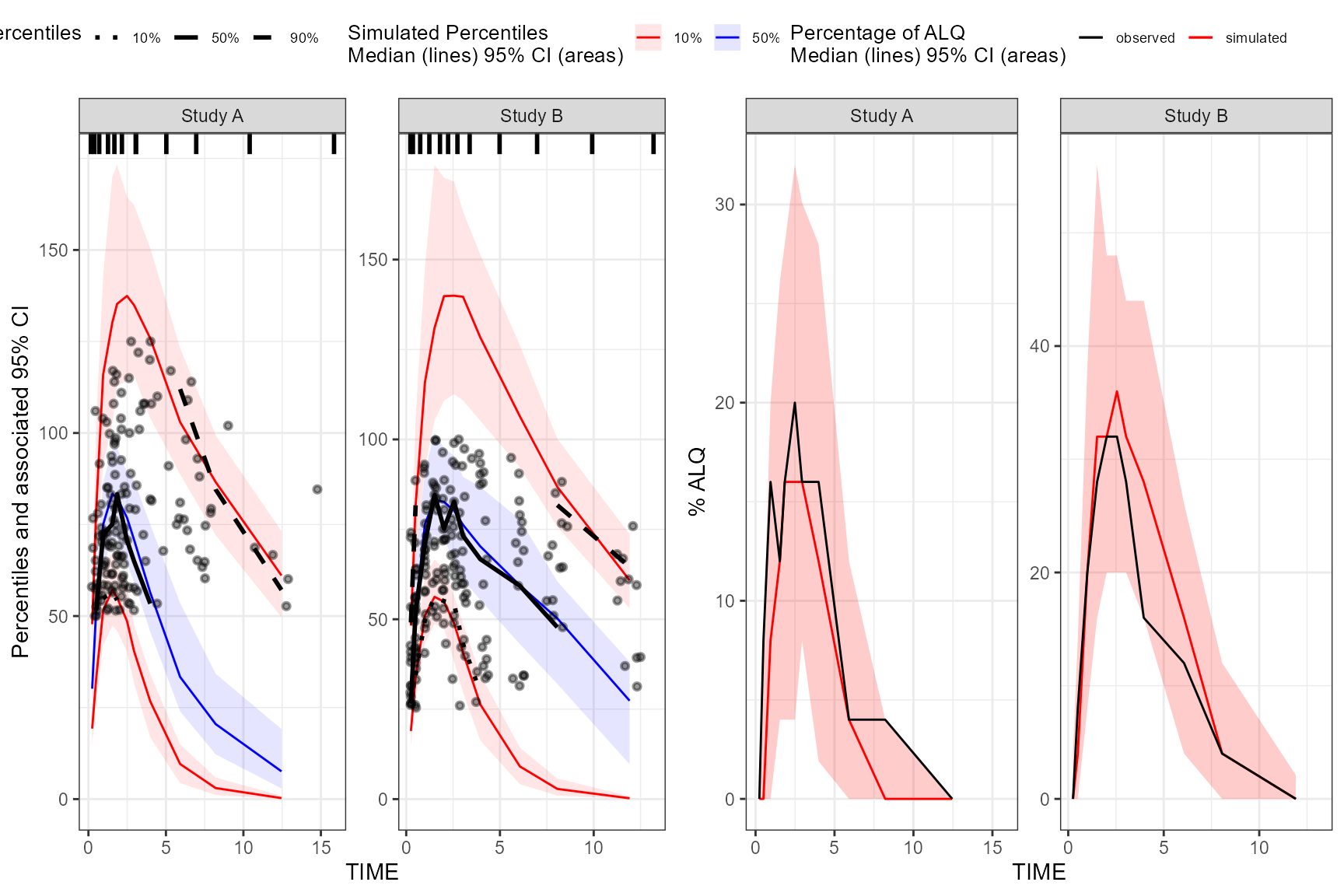
If both blq and alq data, users may supply
censoring.type = "both"
vpc_plots <- plot(vpc, censoring.type = "both", censoring.output = "list")By default, when censoring.tidyvpcobj is used, no
percentage blq/alq plots will be returned e.g., default for
censoring.type = 'none'. If users specify
censoring.type='both' and only blq censoring was performed,
for example, they will receive an error stating e.g.,
pctalq data.frame was not found in tidyvpcobj. Use censoring() to create censored data for plotting alq.
tidyvpc 1.4.0
Additional Binning Methods
The following additional binning methods from classInt
have been made available in tidyvpc.
see ?classIntervals ‘style’ descriptions for applicable
arguments for each selected binning method.
headtails
observed(obs_data, x = TIME, y = DV) |>
simulated(sim_data, y = DV) |>
binning(bin = "headtails") |>
plot()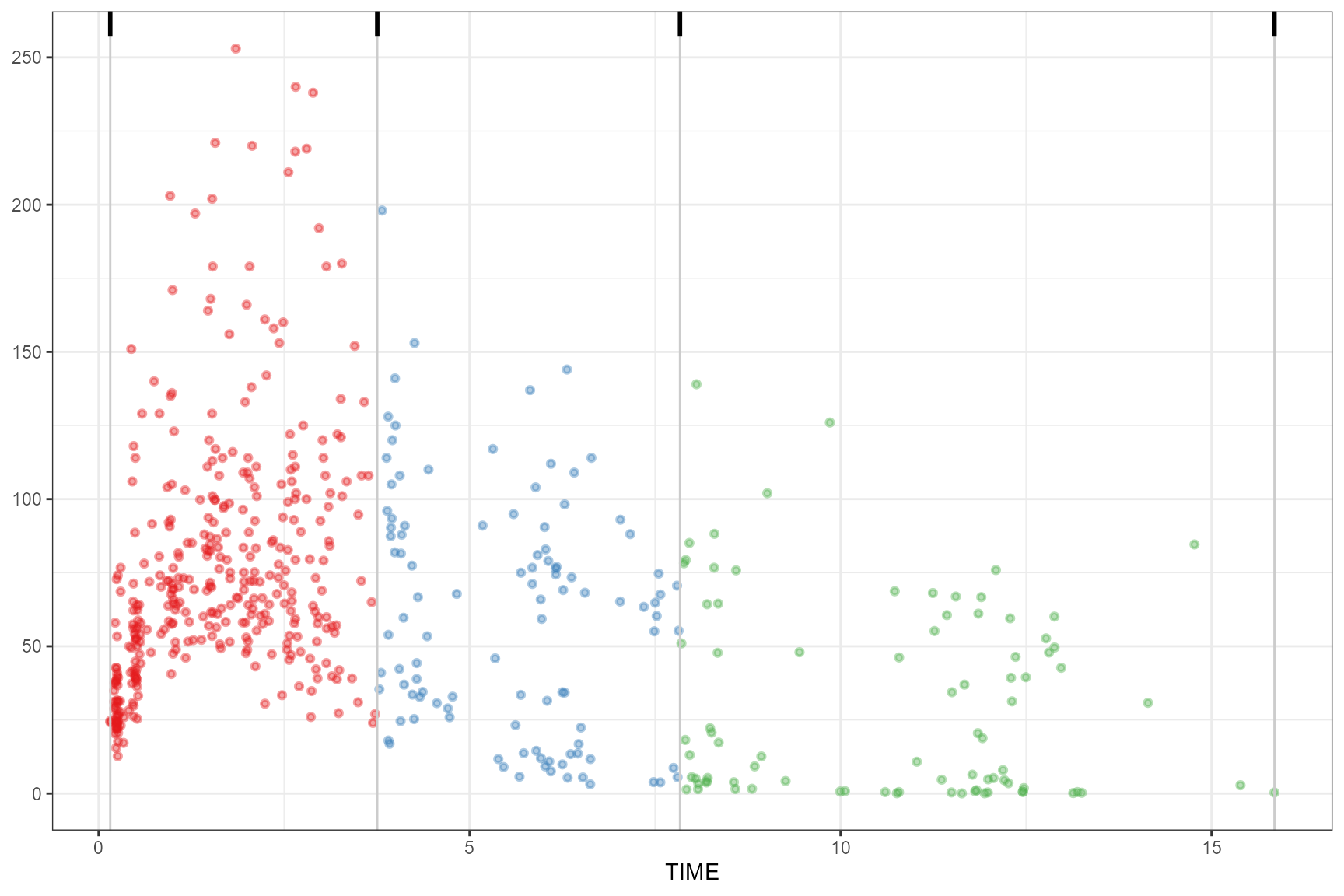
Including additional thr argument.
observed(obs_data, x = TIME, y = DV) |>
simulated(sim_data, y = DV) |>
binning(bin = "headtails", thr = 0.55) |>
plot()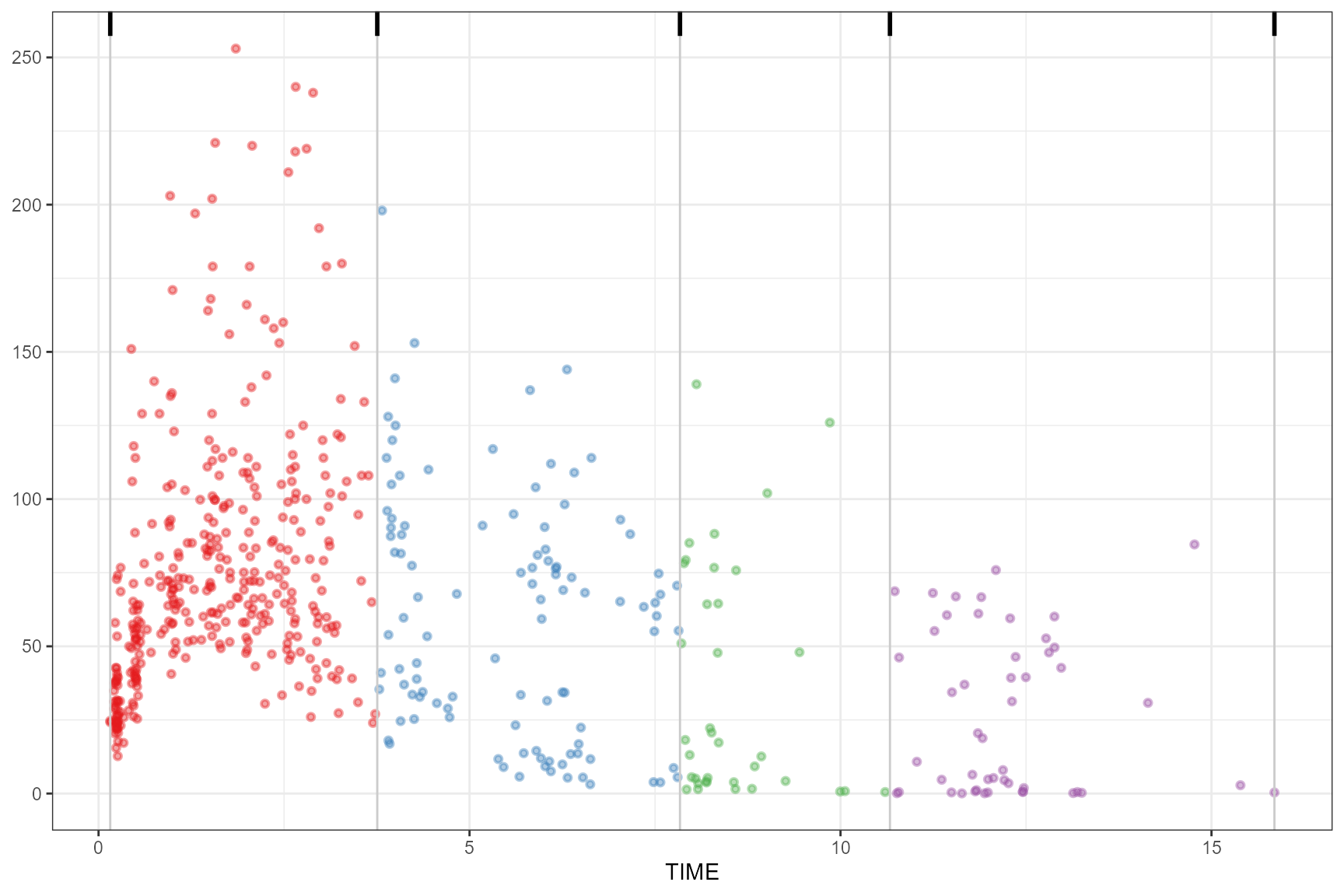
maximum
observed(obs_data, x = TIME, y = DV) |>
simulated(sim_data, y = DV) |>
binning(bin = "maximum") |>
plot()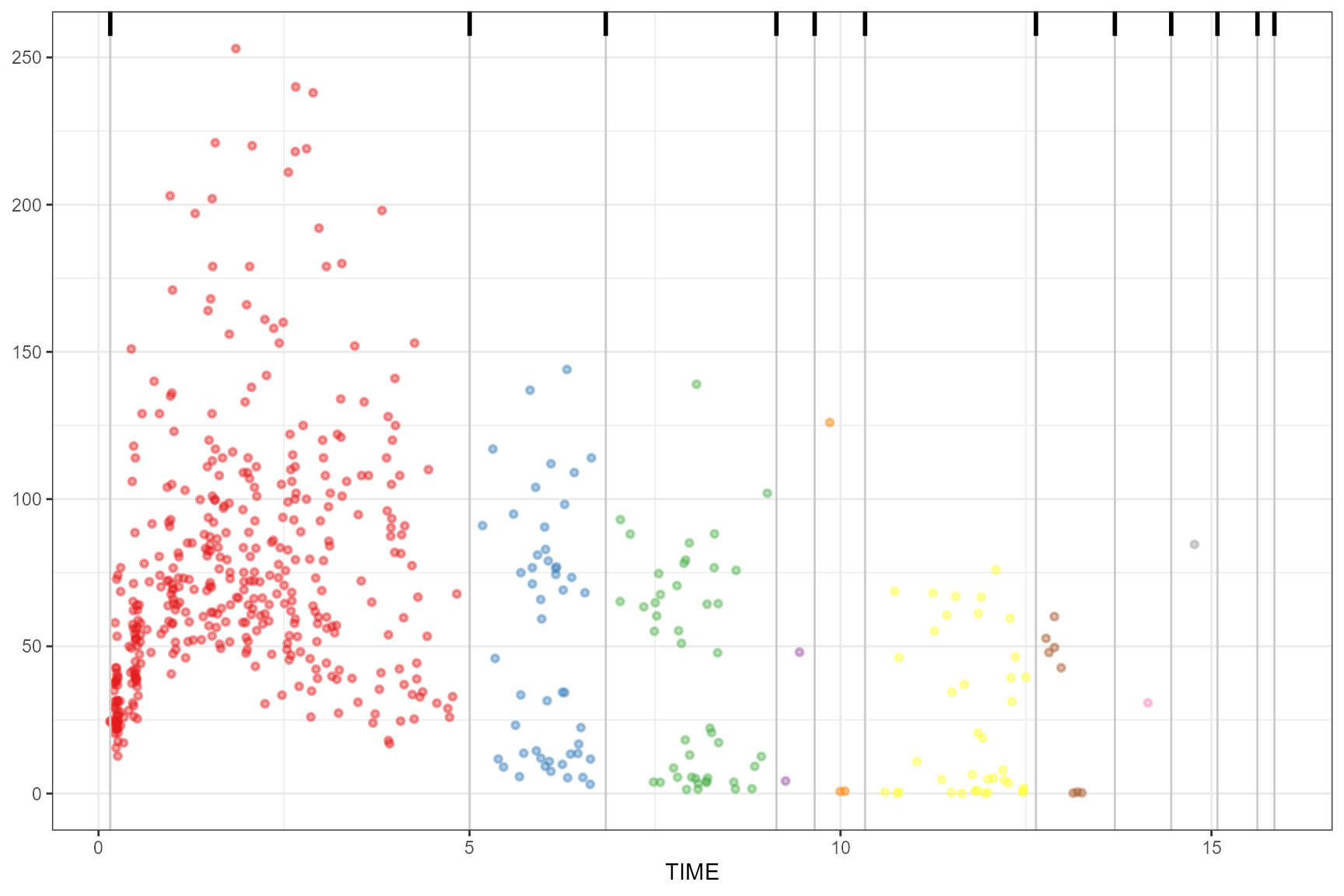
Including additional nbins argument.
observed(obs_data, x = TIME, y = DV) |>
simulated(sim_data, y = DV) |>
binning(bin = "maximum", nbins = 7) |>
plot()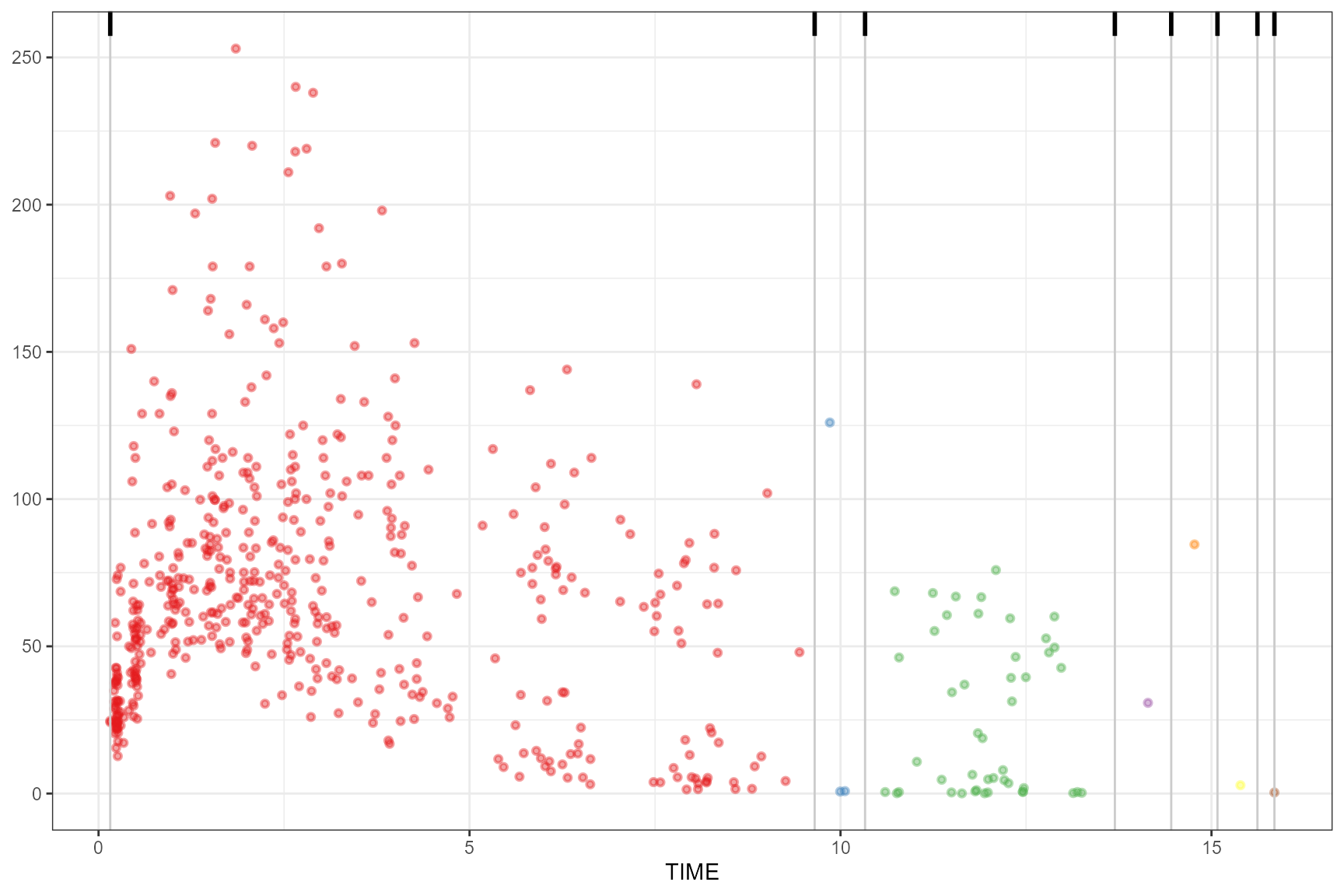
box
observed(obs_data, x = TIME, y = DV) |>
simulated(sim_data, y = DV) |>
binning(bin = "box") |>
plot()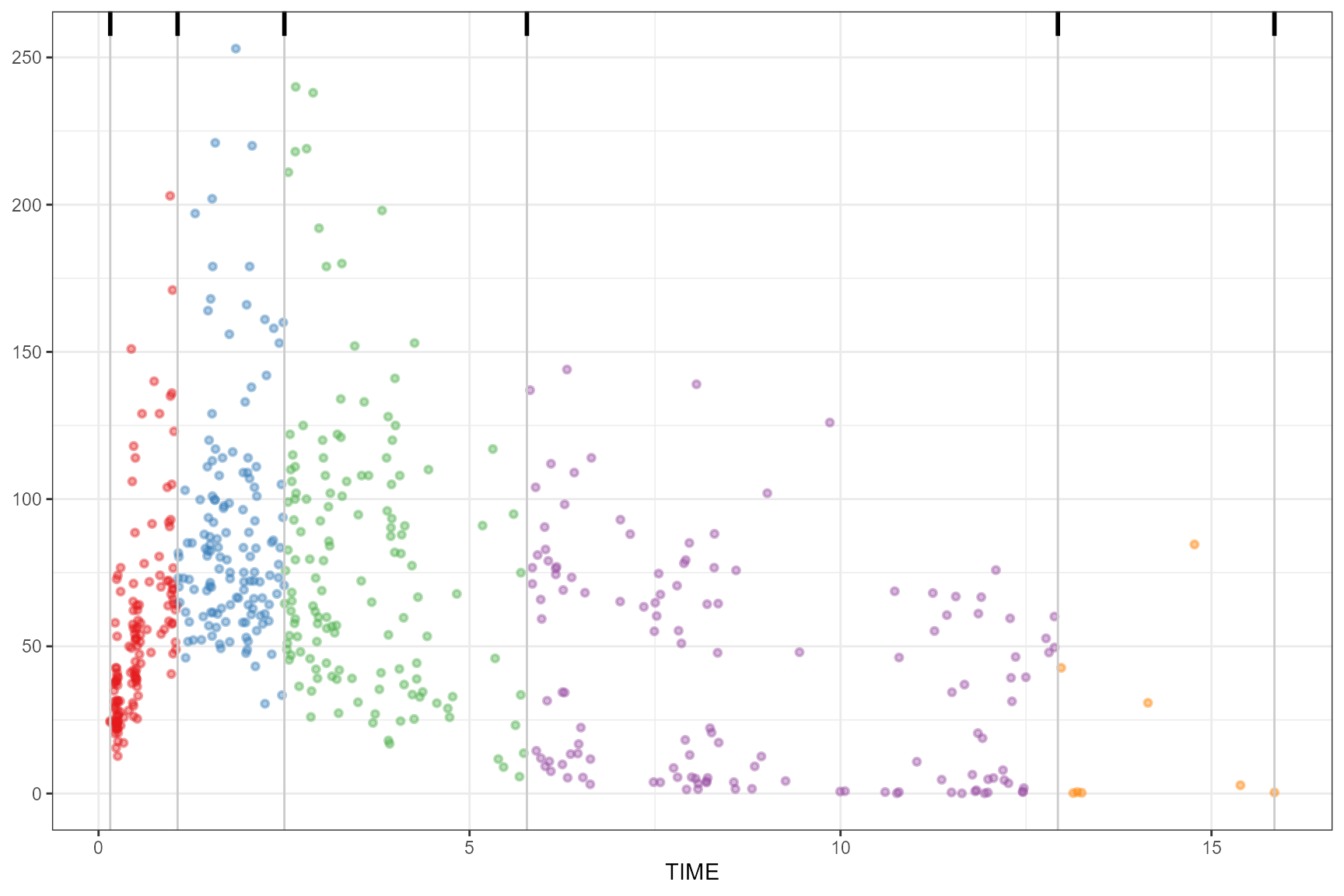
Including additional iqr_mult and type
argument.
observed(obs_data, x = TIME, y = DV) |>
simulated(sim_data, y = DV) |>
binning(bin = "box", iqr_mult = 4) |>
plot()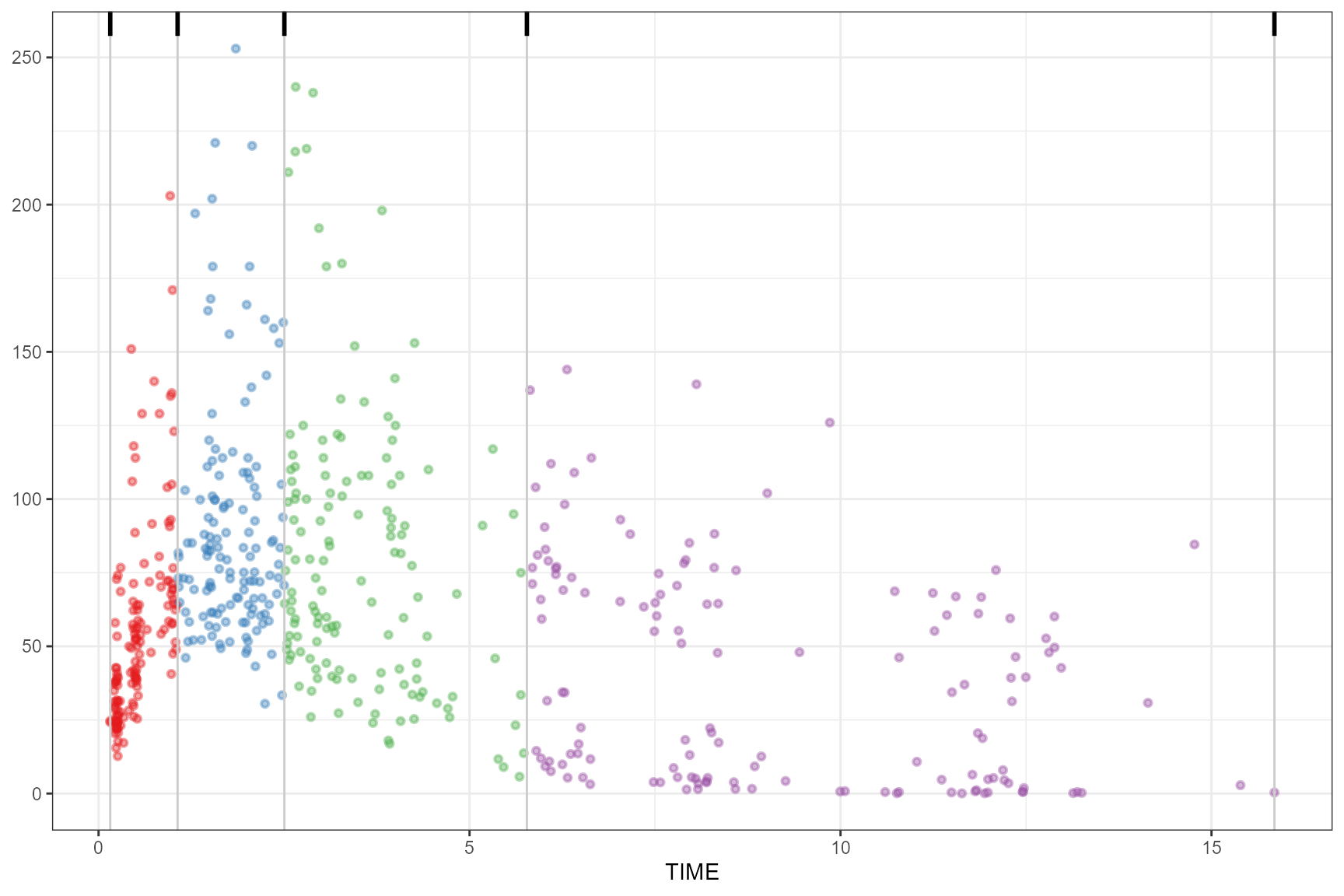
# additional (quantile) type arg
observed(obs_data, x = TIME, y = DV) |>
simulated(sim_data, y = DV) |>
binning(bin = "box", type = 3) |>
plot()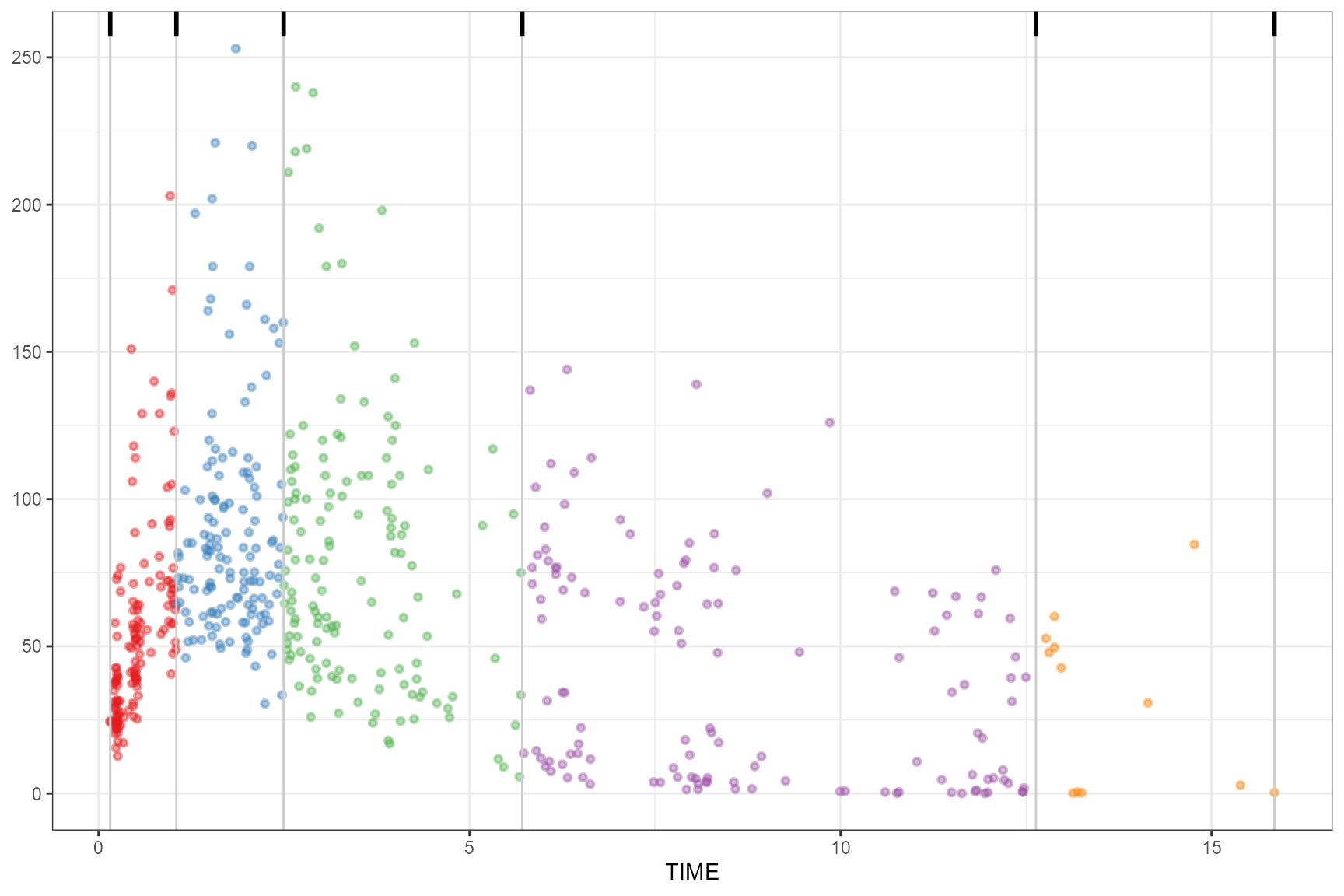
Additional flexibility for binless() + predcorrect()
Users may now execute predcorrect() either before, or
after calling binless(loess.ypc=TRUE). Previously, you were
required to execute predcorrect() before
binless(loess.ypc=TRUE), otherwise you’d receive an
error.
The following code below produces equivalent output:
observed(obs_data, x = TIME, y = DV ) |>
simulated(sim_data, y = DV) |>
stratify(~ GENDER) |>
predcorrect(pred=PRED) |> #before binless()
binless(loess.ypc=TRUE) |>
vpcstats() |>
plot()
observed(obs_data, x = TIME, y = DV ) |>
simulated(sim_data, y = DV) |>
stratify(~ GENDER) |>
binless(loess.ypc=TRUE) |>
predcorrect(pred=PRED) |> #after binless()
vpcstats() |>
plot()tidyvpc 1.3.0
An overview of updates to plot() function in
tidyvpc v1.3.0
Set plot output dimensions:
knitr::opts_chunk$set(fig.width=12, fig.height=8, dpi = 300) One sided stratify() formula uses facet_wrap()
library(tidyvpc)
library(magrittr)
obs_data <- obs_data[MDV == 0]
sim_data <- sim_data[MDV == 0]
vpc <- observed(obs_data, x=TIME, y=DV) %>%
simulated(sim_data, y=DV) %>%
stratify(~ GENDER) %>%
binless() %>%
vpcstats()
plot(vpc)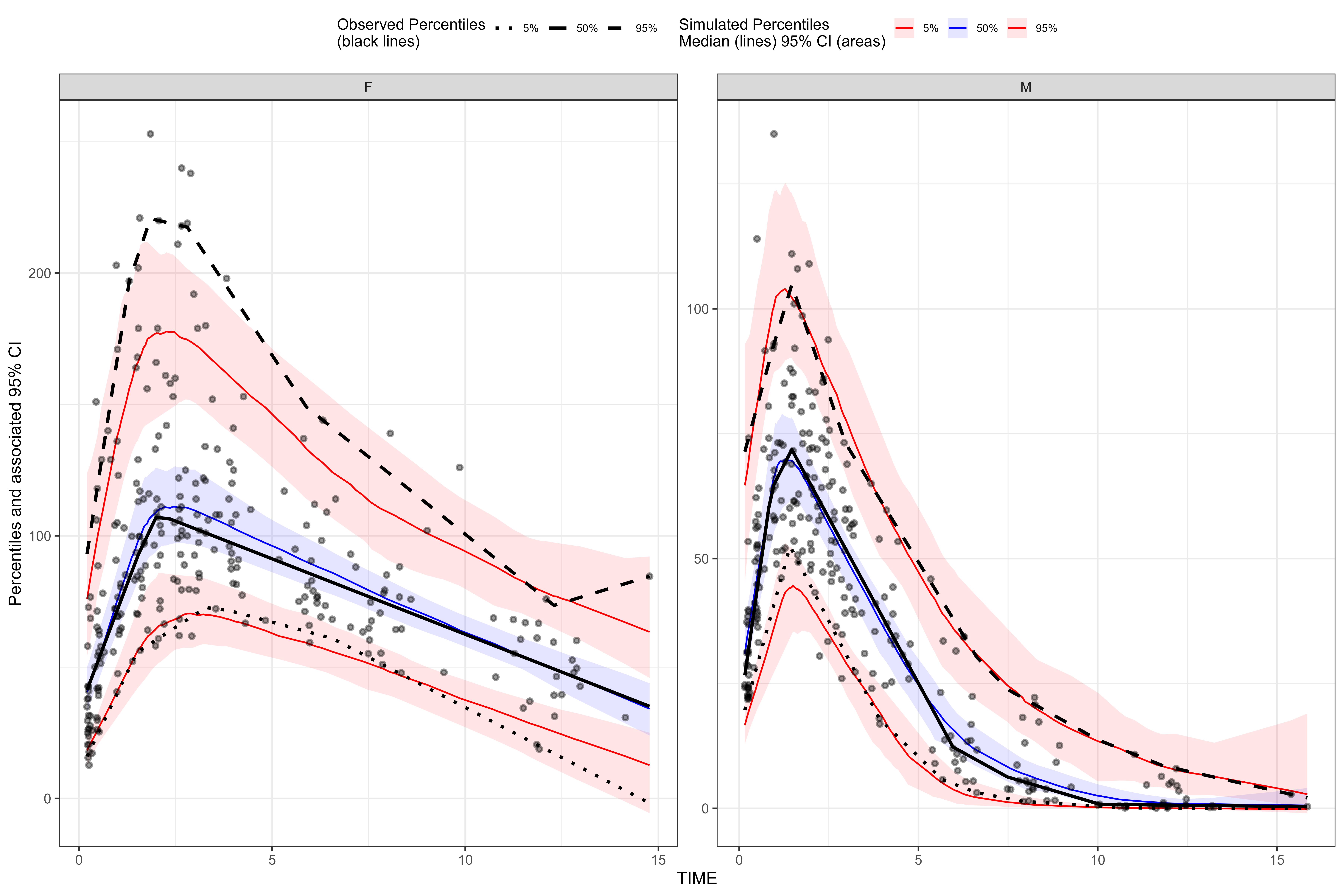
vpc <- observed(obs_data, x=TIME, y=DV) %>%
simulated(sim_data, y=DV) %>%
stratify(~ GENDER + STUDY) %>%
binning(bin = "jenks", nbins = 8) %>%
vpcstats()
plot(vpc)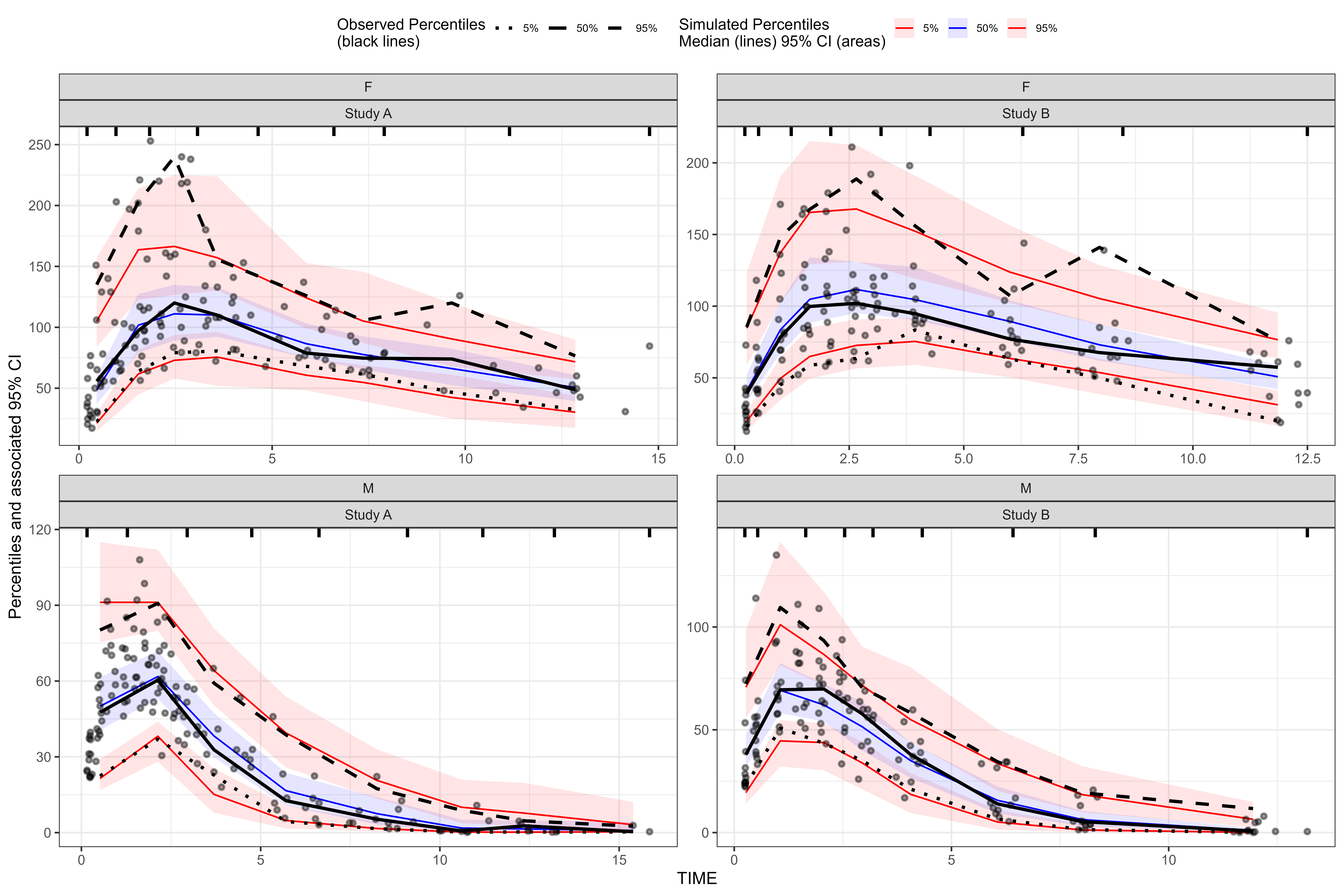
Two-sided formula uses facet_grid()
vpc <- observed(obs_data, x=TIME, y=DV) %>%
simulated(sim_data, y=DV) %>%
stratify(GENDER ~ STUDY) %>%
binning(bin = "kmeans", nbins = 6) %>%
vpcstats()
plot(vpc)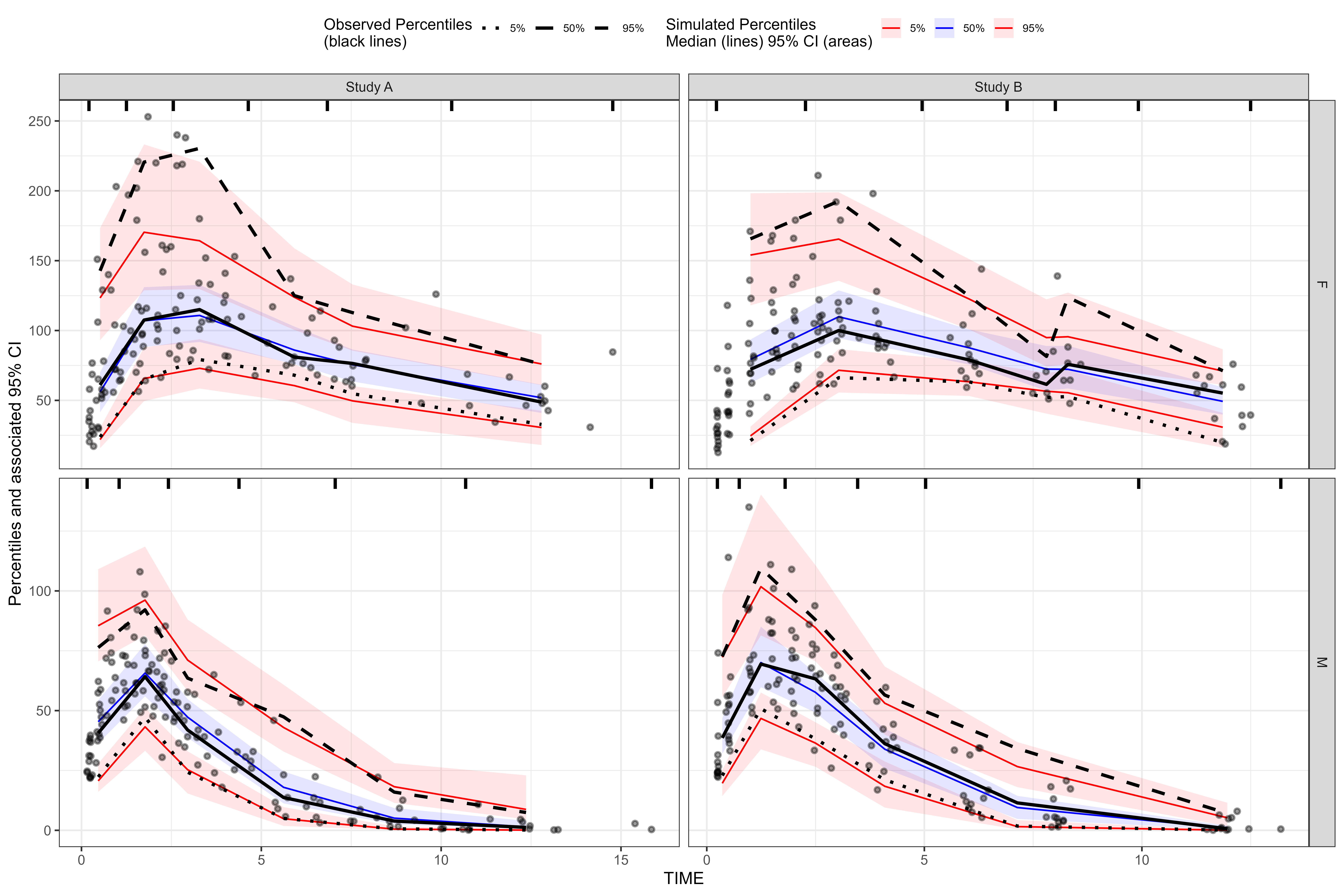
Using facet = TRUE argument
We can use facet = TRUE argument to facet continuous VPC by quantile or facet categorical VPC by predicted probability.
vpc <- observed(obs_data, x=TIME, y=DV) %>%
simulated(sim_data, y=DV) %>%
binless() %>%
vpcstats()
plot(vpc, facet = TRUE, point.alpha = 0.1, point.size = 1, ribbon.alpha = 0.2)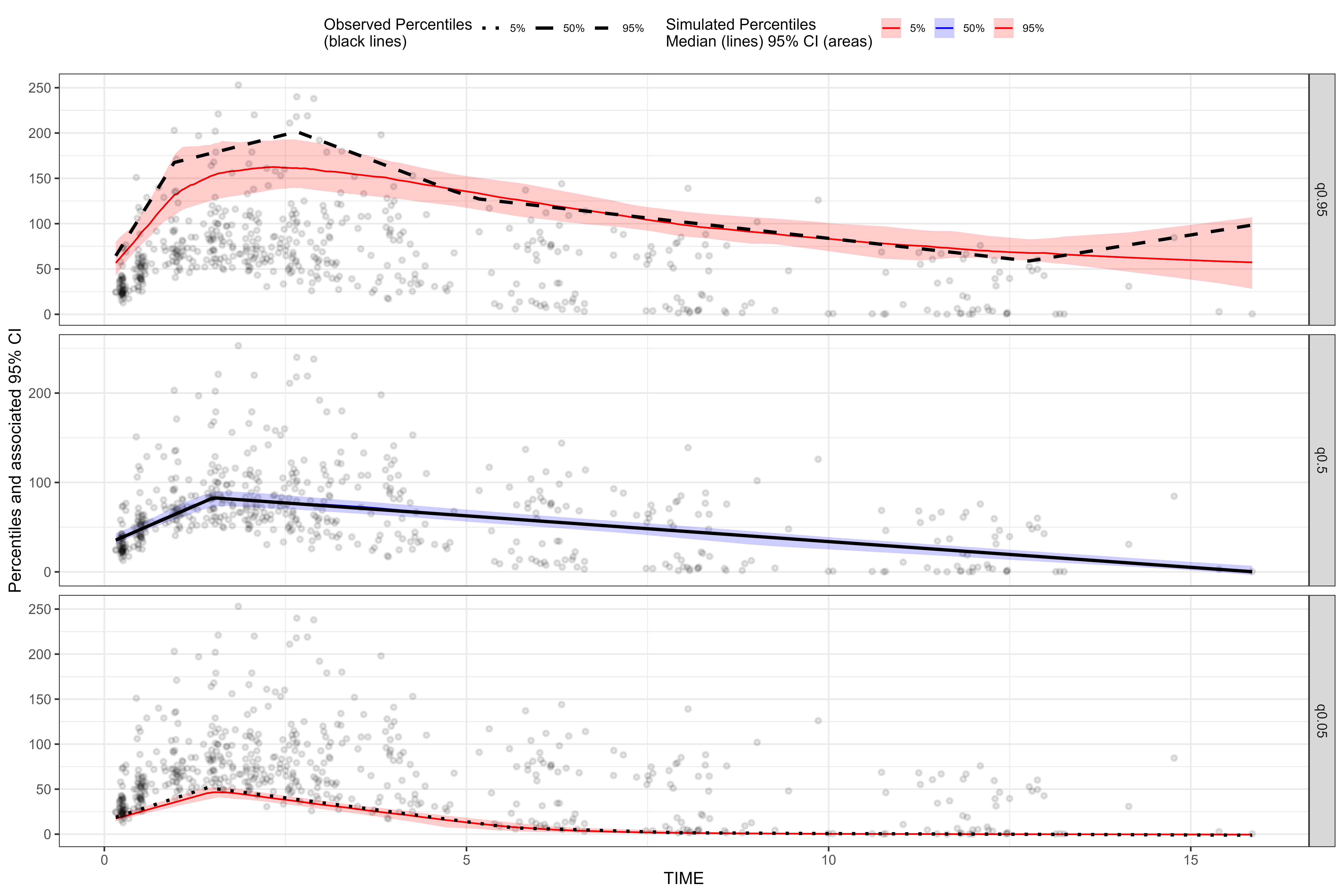
vpc <- observed(obs_cat_data, x = agemonths, yobs = zlencat) %>%
simulated(sim_cat_data, ysim = DV) %>%
binless() %>%
vpcstats(vpc.type = "categorical")
plot(vpc, facet = TRUE, legend.position = "bottom")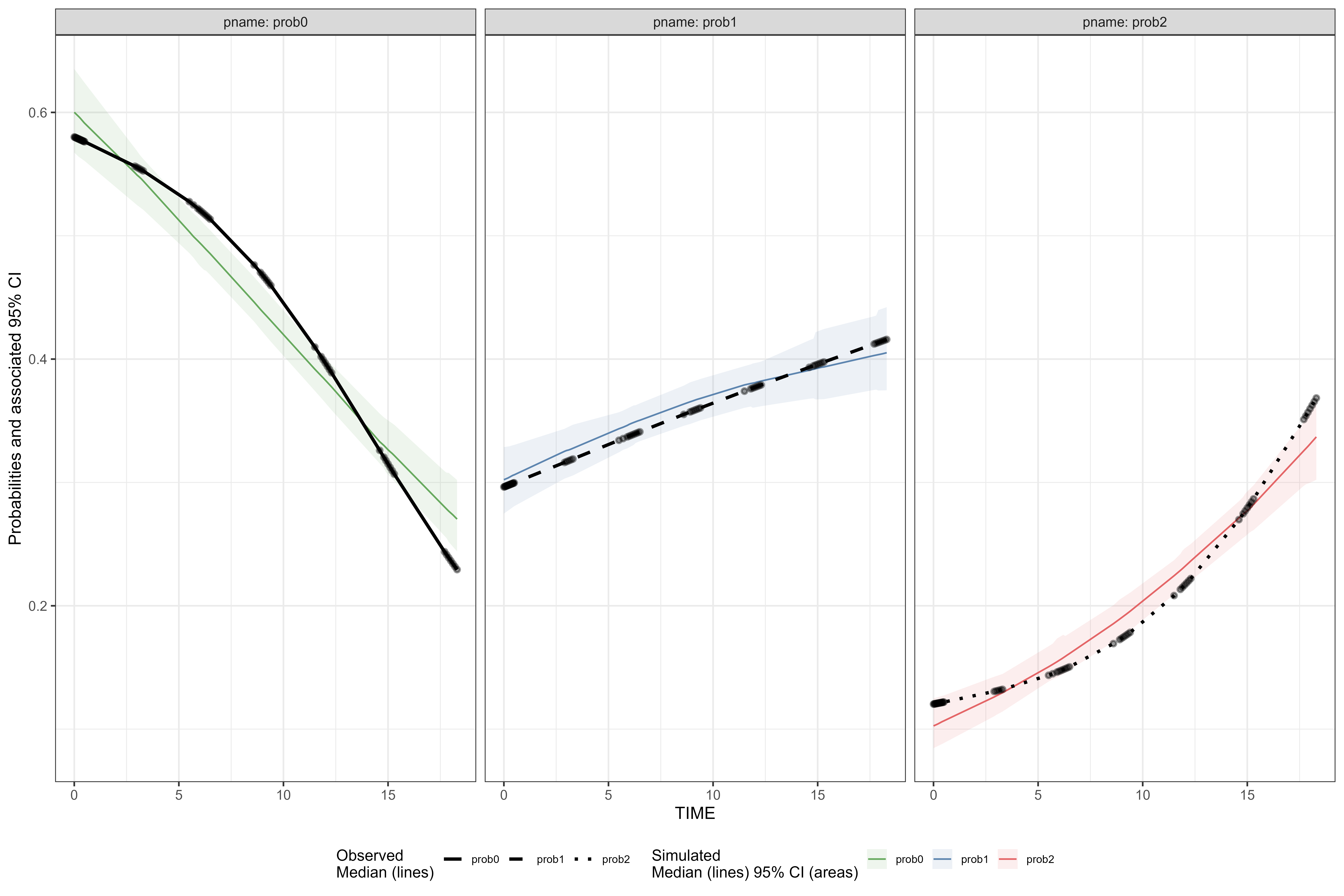
Changing point size, point alpha, point shape, point stroke, and ribbon alpha
Setup categorical VPC.
vpc <- observed(obs_cat_data, x = agemonths, yobs = zlencat) %>%
simulated(sim_cat_data, ysim = DV) %>%
binning(bin = round(agemonths, 0)) %>%
vpcstats(vpc.type = "categorical")Adjust point size.
plot(vpc, point.size = 4)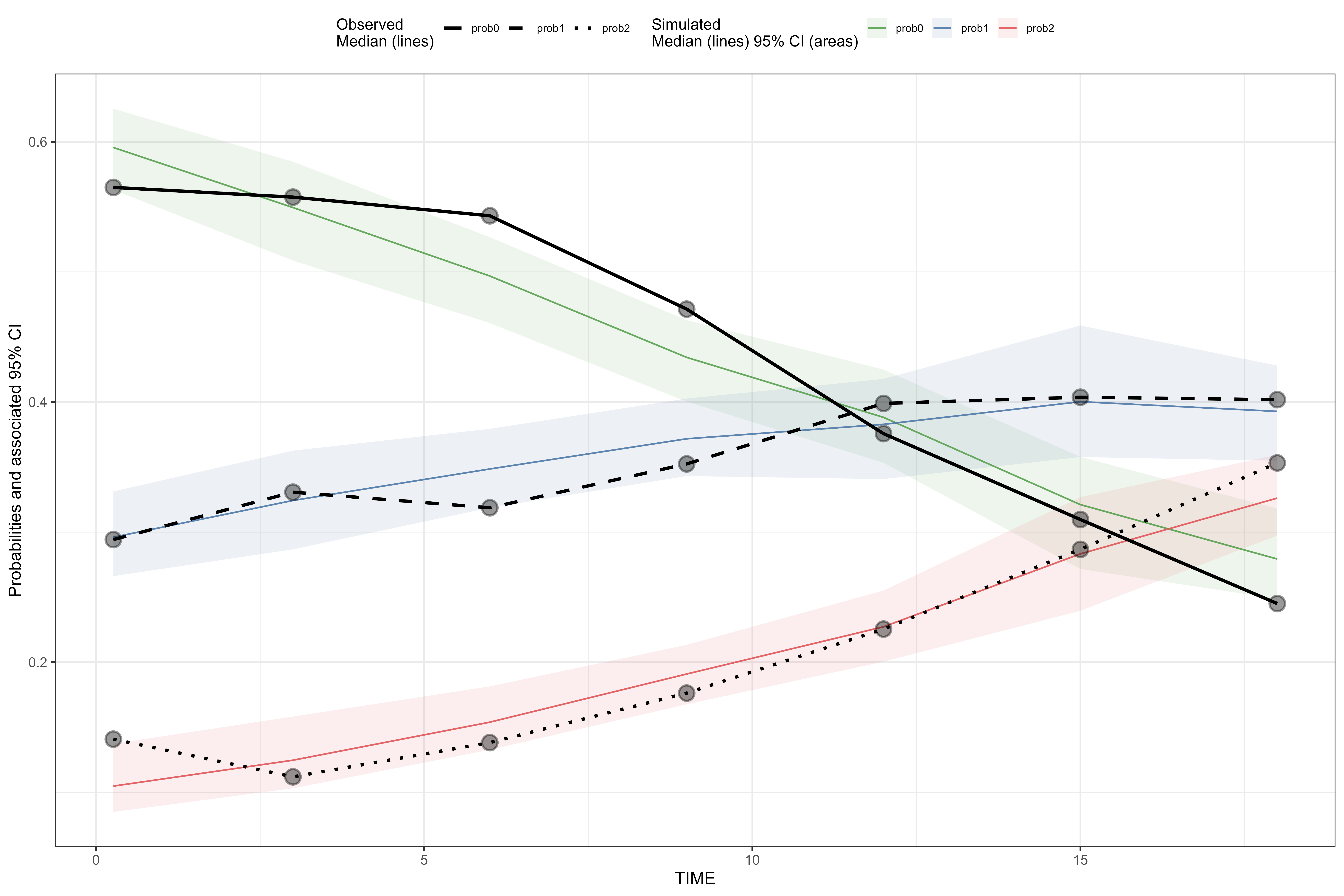
Setup continuous VPC.
plot(vpc, point.size = 1.5, point.stroke = 2.5, point.alpha = 0.1, ribbon.alpha = 0.05)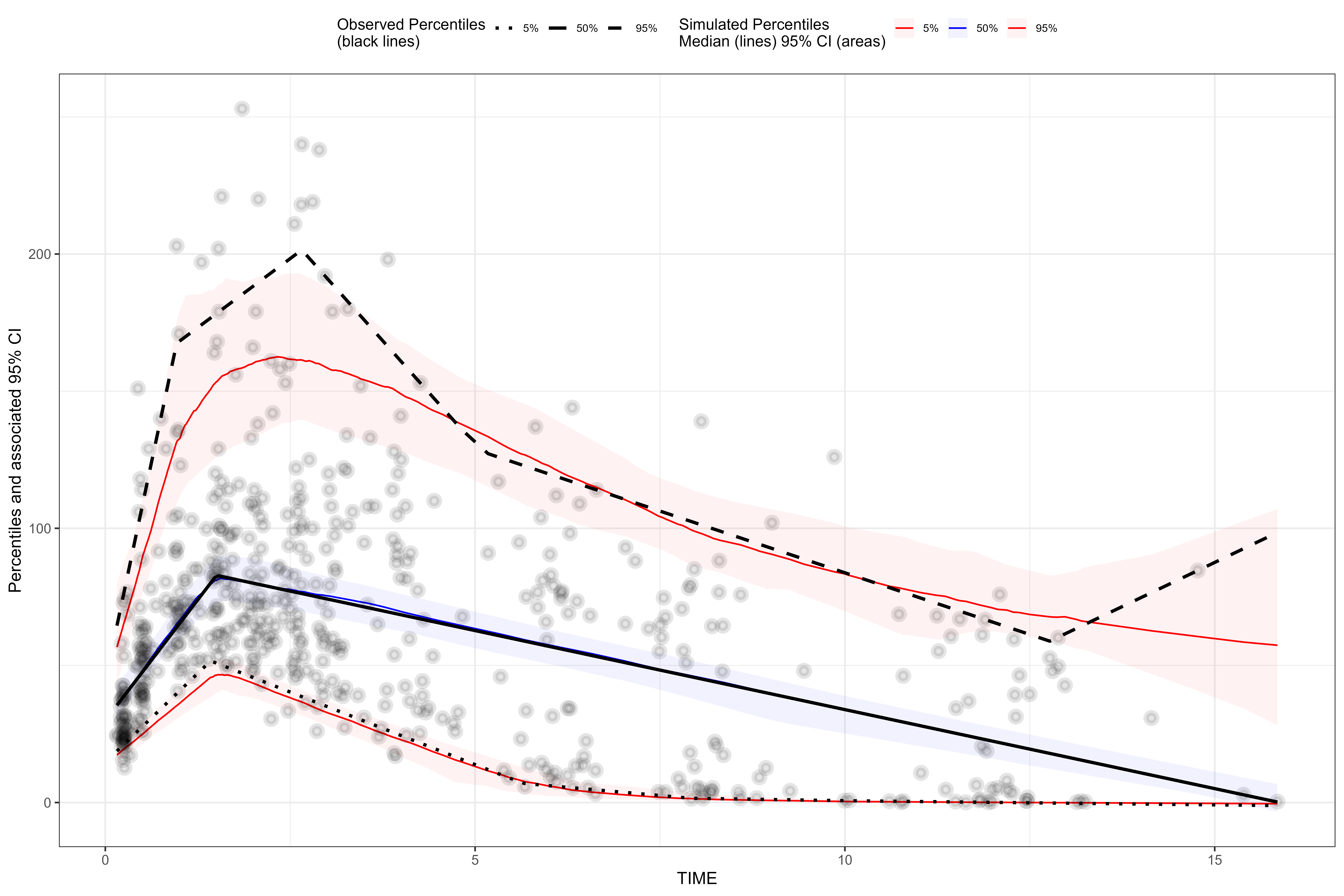
plot(vpc, point.size = 5, point.stroke = 0.3, point.shape = "triangle")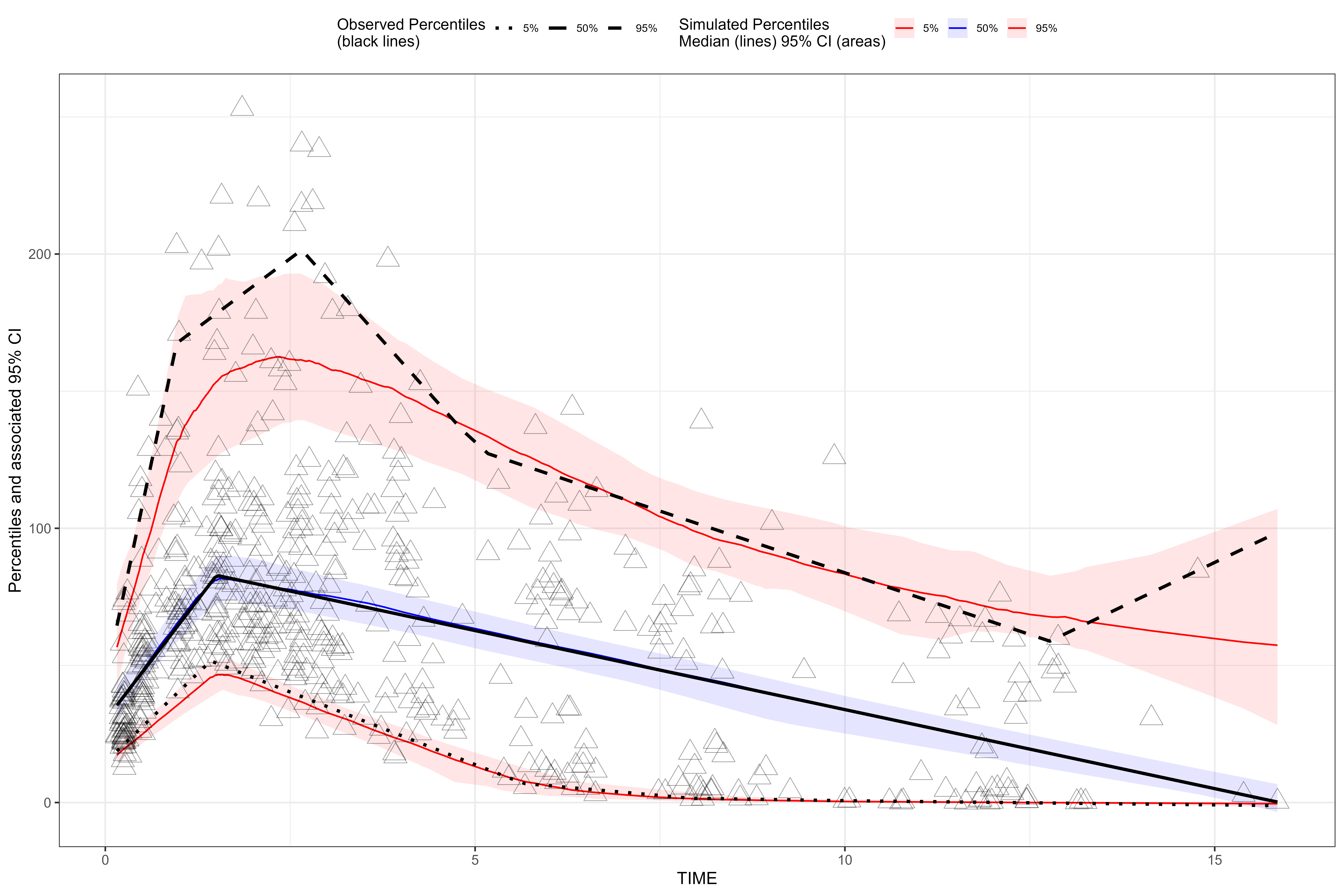
plot(vpc, point.size = 7, point.shape = "square-fill", point.alpha = 0.1, ribbon.alpha = 0.5)