Usage¶
Darwin may be executed locally, on Linux Grids, or as a combination of both (e.g., execute NONMEM models on grids and run search locally).
Execution Overview¶
Running search on local machine¶
The darwin.run_search executes the search for the optimal population model.
python -m darwin.run_search <template_path> <tokens_path> <options_path>
The arguments are the paths to the following files:
Template file (e.g., template.txt) - basic shell for NONMEM control files
Tokens file (e.g., tokens.json) - json file describing the dimensions of the search space and the options in each dimension
Options file (e.g., options.json) - json file describing algorithm, run options, and post-run penalty code configurations.
See Required Files for additional details.
Alternatively, you can execute darwin.run_search or darwin.run_search_in_folder with
a single argument specifying the path to the folder containing the template.txt, tokens.json, and options.json files:
python -m darwin.run_search <folder_path>
python -m darwin.run_search_in_folder <folder_path>
Note
Files must be named as template.txt, tokens.json, and options.json when using the folder argument.
Stopping Execution¶
A running search can be stopped using the following command:
python -m darwin.stop_search [-f] <project dir>|<options file>
Warning
Don’t force-stop GP during the ask stage. Either wait for it to finish (Done asking in the console output and/or messages.txt) or stop without -f flag.
Note
models.json will contain all model runs finished before interruption.
Execution on Linux Grids¶
The following requirements should be met in order to execute pyDarwin on Linux Grids.
You must have access to the grid system (e.g., you are able to connect to the system via terminal session).
You must make pyDarwin installation available for all grid nodes.
Your search project must be available for all grid nodes as well.
You should be familiar with your grid controller commands (e.g., how to submit a job, query finished jobs, and delete jobs).
You should be familiar with regular expressions e.g., for usage in
submit_job_id_reandpoll_job_id_refields inoptions.json.model_run_man must be set to
darwin.GridRunManager.
Note
If all grid nodes share the same file system, you can simply deploy pyDarwin in your home directory (always use virtual environment!).
There are two ways to utilize grids for search in pyDarwin:
Run search locally, submit individual model runs to the grid (local search, grid model runs).
Submit search to the grid, as well as all the model runs (grid search, grid model runs).
Note
Although it’s possible to submit a “local search with local model runs” to the grid, this is not suggested.
In both cases you can stop the search using darwin.stop_search. Just keep in mind that in the second case, it may not be very responsive (due to load/IO latency/grid deployment details), so be patient.
With either case, you need to setup grid settings in your options.json. Below are examples for different workload managers:
"model_run_man": "darwin.GridRunManager",
"generic_grid_adapter": {
"python_path": "~/darwin/venv/bin/python",
"submit_job_id_re": "Your job (\\w+) \\(\".+?\"\\) has been submitted",
"poll_job_id_re": "^\\s+(\\w+)",
"poll_interval": 10,
"poll_command": "qstat -s z",
"delete_command": "qdel {project_stem}-*",
"submit_command": "qsub -b y -o {results_dir}/{run_name}.out -e {results_dir}/{run_name}.err -N {job_name}",
"submit_search_command": "qsub -b y -wd {project_dir} -o {project_stem}.out -e {project_stem}.err -N '{project_stem}'"
},
"model_run_man": "darwin.GridRunManager",
"generic_grid_adapter": {
"python_path": "~/darwin/venv/bin/python",
"submit_job_id_re": "Submitted batch job (\\d+)",
"poll_job_id_re": "^(\\d+)",
"poll_interval": 10,
"poll_command": "squeue -t CD,CA,F,ST,TO --noheader --format=%i",
"delete_command": "scancel {job_ids}",
"submit_command": "sbatch --job-name {job_name} --output {results_dir}/{run_name}.out --error {results_dir}/{run_name}.err --wrap {darwin_cmd}",
"submit_search_command": "sbatch -D {project_dir} --job-name '{project_name}' --output {project_stem}.out --error {project_stem}.err --wrap '{darwin_cmd}'"
},
"model_run_man": "darwin.GridRunManager",
"generic_grid_adapter": {
"python_path": "~/darwin/venv/bin/python",
"submit_job_id_re": "^(\\d+)\\.",
"poll_job_id_re": "^(\\d+)\\.",
"poll_interval": 10,
"poll_command": "qstat -a | grep \"\\b [CE] \"",
"delete_command": "qdel {job_ids}",
"submit_command": "jsub -o {results_dir}/{run_name}.out -e {results_dir}/{run_name}.err -N {job_name} --",
"submit_search_command": "jsub -d {project_dir} -o {project_stem}.out -e {project_stem}.err -N '{project_stem}' --"
},
"model_run_man": "darwin.GridRunManager",
"generic_grid_adapter": {
"python_path": "~/darwin/venv/bin/python",
"submit_job_id_re": "Job \\<(\\d+)\\> is submitted",
"poll_job_id_re": "^(\\d+)",
"poll_interval": 20,
"poll_command": "bjobs -d",
"delete_command": "bkill -J {project_stem}-*",
"submit_command": "bsub -n 1 -q standard -o {results_dir}/{run_name}.out -e {results_dir}/{run_name}.err -J {job_name}",
"submit_search_command": "bsub -q standard -cwd {project_dir} -o {project_stem}.out -e {project_stem}.err -J '{project_stem}'"
},
Running Grid Search¶
You can send the search itself to the grid.
python -m darwin.grid.run_search <template_path> <tokens_path> <options_path>
Or alternatively, run grid search in folder:
python -m darwin.grid.run_search <folder_path>
python -m darwin.grid.run_search_in_folder <folder_path>
Note
Files must be named as template.txt, tokens.json, and options.json when using the folder argument.
Warning
Prior to pyDarwin 3.1.0 either . or absolute path must be used as folder_path.
A few things to do before that:
Ensure that submit_search_command has been set up correctly in options.json, in addition to other grid settings.
Make sure your search job won’t be killed by time-out. The search can take days in some cases, so either set the timeout explicitly in
submit_search_commandor ask your grid administrator for help.Try running the search locally before sending it to the grid: it’s easier to troubleshoot when you have console output.
Customizing Python Environment¶
In some cases, you may need to perform additional actions before you call pyDarwin. For example, if you use modules, you’ll need to load the corresponding module first. You can achieve that with a simple script:
#!/bin/bash module load pydarwin/3.1.0 python "$@"
Use the script as python_path:
"generic_grid_adapter": { "python_path": "~/darwin/run_pydarwin.sh", },
This should work for both the model runs and the search (submit_command and submit_search_command).
Search Info¶
python -m darwin.search_info <folder_path>
This command loads the search folder and shows the summary that looks like this:
[01:11:10] Changing directory to c:\workspace\fruitfly\examples\NONMEM\user\Example2
[01:11:10] Options file found at options.json
[01:11:10] Loading system options: c:\workspace\fruitfly\examples\user\options.json
[01:11:10] Template file found at template.txt
[01:11:10] Tokens file found at tokens.json
[01:11:10] Algorithm: GP
[01:11:10] Engine: NONMEM
[01:11:10] random_seed: 11
[01:11:10] Project dir: c:\workspace\fruitfly\examples\NONMEM\user\Example2
[01:11:10] Data dir: c:\workspace\fruitfly\examples\NONMEM\user\Example2
[01:11:10] Project working dir: C:\Users\jcook\pydarwin\Example2
[01:11:10] Project temp dir: C:\Users\jcook\pydarwin\Example2\temp
[01:11:10] Project output dir: C:\Users\jcook\pydarwin\Example2\output
[01:11:10] Key models dir: C:\Users\jcook\pydarwin\Example2\key_models
[01:11:10] Search space size: 12960
[01:11:10] Estimated number of models to run: 454
Required Files¶
The same 3 files are required for any search, whether EX, GA, GP, RF, GBRT, or PSO. Which algorithm is used is defined in the options file. The template file serves as a framework and looks similar to a NONMEM/NMTRAN control file. The tokens file specifies the range of “features” to be searched, and the options file specifies the algorithm, the fitness function, any R or Python code to be executed after the NONMEM execution, and other options related to execution. See Options List.
Template File¶
The template file is a plain ASCII text file. This file is the framework for the construction of the NONMEM control files. Typically, the structure will be quite similar to a NONMEM control file, with the usual blocks, e.g., $PROB, $INPUT, $DATA, $SUBS, $PK, $ERROR, $THETA, $OMEGA, $SIGMA, $EST. However, this format is completely flexible and entire blocks may be missing from the template file (to be provided from the tokens file).
Note
NONMEM does not allow the data set path ($DATA) to be longer than 80 characters and the path must be in quotes if it contains spaces (see Data Directory).
The difference between a standard NONMEM control file and the template file is that the user will define code segments in the template file that will be replaced by other text. These code segments are referred to as “token keys”. Token keys come in sets, and in most cases, several token keys will need to be replaced together to generate syntactically correct code. The syntax for a token key in the template file is:
{Token_stem[N]}
Where Token_stem is a unique identifier for that token set and N is the target text to be substituted. An example is instructive.
Example:
Assume the user would like to consider 1 compartment (ADVAN1) or 2 compartment (ADVAN3) models as a dimension of the search. The relevant template file for this might be:
$SUBS {ADVAN[1]}
.
.
$PK
.
.
.
{ADVAN[2]}
.
.
.
$THETA
(0,1) ; Volume - fixed THETA - always appears
(0,1) ; Clearance - fixed THETA - always appears
{ADVAN[3]}
Note that tokens nearly always come in sets. As in nearly all cases, several substitutions must be made to create correct syntax. For a one compartment model, the following substitutions would be made:
{ADVAN[1]} -> ADVAN1
{ADVAN[2]} -> ;; 1 compartment, no definition needed for K12 or K21
{ADVAN[3]} -> ;; 1 compartment, no initial estimate needed for K12 or K21
and for 2 compartment:
{ADVAN[1]} -> ADVAN3
{ADVAN[2]} -> K12 = THETA(ADVANA) ;; 2 compartment, need definition for K12 \n K21 = THETA(ADVANB)
ADVAN[3]} ->(0,0.5) ;; K12 THETA(ADVANA) \n (0,0.5) ;; K21 THETA(ADVANB)
Where \n is the new line character. These sets of tokens are called token sets (2 token sets in this example one for ADVAN1, one for ADVAN3). The group of token sets is called a token group. In this example, “ADVAN” is the token key. Each token group must have a unique token key. For the first set of options, the text “ADVAN1” is referred to as the token text. Each token set consists of key-text pairs: token keys (described above) and token text.
The token (consisting of “{” + token stem +[N] + “}” where N is an integer specifying which token text in the token set is to be substituted) in the template file is replaced by the token text, specified in the tokens file. Which set of token key-text pairs is substituted is determined by the search algorithm and provided in the phenotype.
Note that the THETA (and ETA and EPS) indices cannot be determined until the final control file is defined, as THETAs may be included in one token set, but missing in another token set. For this reason, all fixed initial estimates in the $THETA block MUST occur before the THETA values that are not fixed (e.g., are searched). This is so the algorithm can parse the resulting file and correctly calculate the appropriate THETA (and ETA and EPS) indices. Further, the text string index in the token (e.g., ADVANA and ADVANB) must be unique in the token groups. The most convenient way to ensure that the text string index is unique in the Token groups is to use the token stem as the THETA index (e.g., THETA(ADVAN) is the token stem is ADVAN). Additional characters (e.g., ADVANA, ADVANB) can be added if multiple THETA text indices are needed. Note that the permitted syntax for residual error is EPS() or ERR().
Special notes on structure of $THETA/$OMEGA/$SIGMA:¶
Parameter initial estimate blocks require special treatment. A template file will typically include 2 types of initial estimates:
Fixed initial estimates - Initial estimates that are not searched, but will be copied from the template into ALL control files. These are the typical $THETA estimates, e.g.: (0,1) ; THETA(1) Clearance.
Searched initial estimates - Initial estimates that are specified in tokens that may or may not be in any given control file, e.g., {ALAG[2]} where the text for the ALAG[2] token key is “(0,1) ;; THETA(ALAG) Absorption lag time”
Note
Fixed initial estimates MUST be placed before searched initial estimates
pyDarwin automatically determines the correct indices for any THETA/OMEGA/SIGMA elements that are part of the search options. In order to correctly number these, it first must determine the number of “fixed” (i.e., present in all models, not searched) elements for each. For this reason, the fixed elements of THETA/OMEGA/SIGMA must come before any searched elements in the $THETA/$OMEGA/$SIGMA blocks. pyDarwin counts the number of fixed initial estimates in (for example) the $THETA block, then starts numbering the searched THETAs with the next consecutive number. As is the case for NONMEM, the indices for fixed THETAs are determined entirely by their position in the $THETA block. That is, if the $THETA block is:
$THETA
0.1
0.1
0.1
These will be THETA(1) to THETA(3) and pyDarwin will start with an index of 4. Errors would occur if pyDarwin simply counted the number THETA/ETA/EPS values in the $PK, $ERROR, $MIX, $AES and $DES block if, for example, fixed THETAs were used in $THETA and they did not appear in $PK. Therefore, the sequencing of THETA/ETA/EPS indices is based on the values and positions in the initial values blocks. In addition, to correctly number the elements, pyDarwin needs a little more help finding the correct indices. Specifically, comments (text after a ‘;’) in the $THETA block and THETA initial estimate tokens MUST be used. If this text is not present, and more than one initial estimate was used in a token set (e.g., CL=THETA(VMAX)*CONC/(THETA(KM)+CONC), pyDarwin would not know which initial estimate is to be associated with THETA(EMAX) and which with THETA(KM).
Generally, it is less confusing to have a separate line for each initial estimate, with a comment for that initial estimate. However, multiple initial estimates can be put on a single line, with multiple ‘;’ separating the defining text (please ensure that you are following naming conventions outlined below).
Specifically, there are 3 ways to define a variable name (ETA/THETA/OMEGA):
; NAME (any amount of spaces before and after name)
; any text ETA(NAME) any text (no spaces between ETA and name or around name)
; any text ETA <on|ON> NAME any text (exactly one space between the words)
Any combination of those in one line must work:
<some complex definition> ; name1 ; name2 ; also ETA(name3) ; be aware that numbers count as well: ETA(4)
Here we have 4 variables (ETAs) which can be referred to as ETA 1 to 4. Keep in mind that every ETA(name) in the model text is replaced with ETA(<number>), even inside the definition block.
If this is not what you want, you may define it using another notation, or add something to the comment:
D = ETA(D1)*ETA(C)*ETA(A)*ETA(D2)
$OMEGA
0.1 ; ETA(D1)
0.1 ; A
0.1 ; ETA ON C
0.1 ; ETA(D2) D2 or ETA ON D2 or any other way that doesn't look like another definition
Which then becomes:
D = ETA(1)*ETA(3)*ETA(2)*ETA(4)
$OMEGA
0.1 ; ETA(1)
0.1 ; A
0.1 ; ETA ON C
0.1 ; ETA(4) D2 or ETA ON D2
Parenthesis with (lower bound, initial value, upper bound) may also be used, as illustrated below:
$PK
D = THETA(1)*THETA(3)*THETA(2)*THETA(4)
$OMEGA
(0,0.1,10) ; THETA(1)
(0,0.1) ; THETA(A)
(0.1) ; THETA(3)
0.1 ; THETA(4)
Tokens File¶
The tokens file provides a dictionary (as a JSON file) of token key-text pairs. The highest level of the dictionary is the token group. Token groups are defined by a unique token stem. The token stem also typically serves as the key in the token key-text pairs. The token stem is a text string that corresponds to the token key that appears in the template file. The 2nd level in the tokens dictionary is the token sets. In the template file the tokens are indexed (e.g., ADVAN[1]), as typically multiple token keys will need to be replaced by text to create correct syntax. For example, if the search is for 1 compartment (ADVAN1) vs 2 compartment (ADVAN3), for ADVAN3, definitions of K23 and K32 must be provided in the $PK block, and (typically) initial estimates must be provided in the $THETA block. Thus, a set of 3 replacements must be made, one in $SUBS, one in $PK, and one in $THETA. So, the token set for selection of number of compartments, for 1 compartment (first option) or 2 compartments (second option), will include the following JSON code:
"ADVAN": [
["ADVAN1 ",
";; 1 compartment, no definition needed for K12 or K21 ",
";; 1 compartment, no initial estimate needed for K12 or K21"
],
["ADVAN3 ",
" K12 = THETA(ADVANA) ;; 2 compartment, need definition for K12 \n K21 = THETA(ADVANB)",
" (0,0.5) ;; K12 THETA(ADVANA) \n (0,0.5) ;; K21 THETA(ADVANB) "
],
Note that specification of the current model as one compartment or two is done by the search algorithm and provided in the model phenotype.
A diagram of the token structure is given below
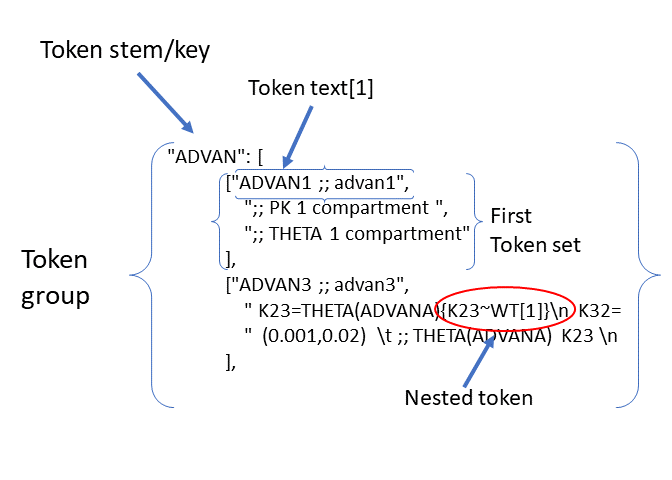
Note the “nested token” – a token (“{K23~WT[1]}”) within a token, circled in red. Any number of levels of nested tokens is permitted (but the logic becomes very difficult with more than one). pyDarwin will first substitute the full text into the template, then scans the resulting text again. This nested token will then be found and the text from the {K23~WT[1]} token set will be substituted.
Several notes:
1. The token stem is “ADVAN” and identifies the token group. This stem must be unique in the token groups. The token stem also typically serves as the token key in the token key-value pairs. In this example, three replacements must be made in the template, in $SUBS, $PK, and $THETA. In the template file, these will be coded as {ADVAN[1]}, {ADVAN[2]}, and {ADVAN[3]}. Note the curly braces, these are required in the template, but not the tokens file. The indices correspond to the indices of the tokens in the token set. In this case there are 3 token key-value pairs in each token set. There may be additional unused tokens (as may be the case with nested tokens) but each token in the template file must have a corresponding token key-value pair in the tokens file. There are 2 token sets in this token group, one coding for ADVAN1 and one coding for ADVAN3.
New lines in JSON files are ignored. To code a new line, enter the newline escape character “\n”. Similarly, a tab is coded as “\t”.
Comments are not permitted in JSON files. However, comments for the generated NMTRAN control file maybe included with the usual syntax “;”.
There is no dependency on the sequence of token sets in the file, any order is acceptable, they need not be in the same order as they appear in the template file.
All other JSON rules apply.
Special note on initial estimates
In order to parse the text in the initial estimates blocks (THETA, OMEGA, and SIGMA) the user MUST include token stem text as a NONMEM/NMTRAN comment (i.e., after “;”). There is no other way to identify which initial estimates are to be associated with which THETA. For example, if a token stem has two THETAs:
Effect = THETA(EMAX) * CONC/(THETA(EC50) + CONC)
for the text in the $PK block, then code to be put into the $THETA block will be:
" (0,100) \t; THETA(EMAX) "
" (0,1000) \t; THETA(EC50) "
Where \t is a tab. Without this THETA(EMAX) and THETA(EC50) as a comment, there would not be any way to identify which initial estimate is to be associated with which THETA. Note that NONMEM assigns THETAs by sequence of appearance in $THETA. Given that the actual indices for THETA cannot be determined until the control file is created, this approach would lead to ambiguity. Each initial estimate must be on a new line and include the THETA (or ETA or EPS) + parameter identifier.
Options File¶
A JSON file with key-value pairs specifying various options for executing pyDarwin. While some fields are mandatory, some are algorithm-specific, while others are only relevant for execution on Linux Grids.
See Options List for details.
Searching Omega Structure¶
In addition to specifying relations inside the template file and tokens file to define the search space, you may also search for different structures of the omega matrix given fields specified in options.json.
Note
Omega structure alone can be searched without any tokens for compartments, covariates, etc. If searching Omega submatrices, options for Omega band/block search must be additionally specified.
Omega structure is encoded by a set of separate genes: one of the genes represents the omega block pattern, another one is for the band width (only applicable to NONMEM models). The pattern is the index of a valid pattern composed by pyDarwin.
In case of independent omega search, the set is repeated as many times as the number of search blocks in the template.
Valid patterns are created based on the maximum omega search block length and maximum size of submatrices (specified through max_omega_sub_matrix, see Omega Submatrices Search for details) if applicable. For example, for search_block(A, B, C, D, E) and max_omega_sub_matrix = 4, pyDarwin will consider the following 16 patterns:
()
(A B C D E)
(A B)
(A B) (C D)
(A B) (C D E)
(A B) (D E)
(A B C)
(A B C) (D E)
(A B C D)
(B C)
(B C) (D E)
(B C D)
(B C D E)
(C D)
(C D E)
(D E)
Here the empty pattern, (), means there is no block Omega (i.e., everything is diagonal), and the variables enclosed by the parenthesis are the ones whose associated covariance matrix (Omega) is block (that is, for each pattern, only those variables whose Omega matrix is block are listed). For NONMEM models without submatrix search, the empty pattern is substituted with an extra value for band width gene (= 0).
The number of patterns for different combinations of max_omega_search_len (whose values listed in the first column) and max_omega_sub_matrix (whose values listed in the first row) can be found in the table below.
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
15 |
16 |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
3 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
6 |
8 |
8 |
8 |
8 |
8 |
8 |
8 |
8 |
8 |
8 |
8 |
8 |
8 |
8 |
5 |
9 |
14 |
16 |
16 |
16 |
16 |
16 |
16 |
16 |
16 |
16 |
16 |
16 |
16 |
16 |
6 |
14 |
25 |
30 |
32 |
32 |
32 |
32 |
32 |
32 |
32 |
32 |
32 |
32 |
32 |
32 |
7 |
22 |
45 |
57 |
62 |
64 |
64 |
64 |
64 |
64 |
64 |
64 |
64 |
64 |
64 |
64 |
8 |
35 |
82 |
109 |
121 |
126 |
128 |
128 |
128 |
128 |
128 |
128 |
128 |
128 |
128 |
128 |
9 |
56 |
150 |
209 |
237 |
249 |
254 |
256 |
256 |
256 |
256 |
256 |
256 |
256 |
256 |
256 |
10 |
90 |
275 |
402 |
465 |
493 |
505 |
510 |
512 |
512 |
512 |
512 |
512 |
512 |
512 |
512 |
11 |
145 |
505 |
774 |
913 |
977 |
1005 |
1017 |
1022 |
1024 |
1024 |
1024 |
1024 |
1024 |
1024 |
1024 |
12 |
234 |
928 |
1491 |
1794 |
1937 |
2001 |
2029 |
2041 |
2046 |
2048 |
2048 |
2048 |
2048 |
2048 |
2048 |
13 |
378 |
1706 |
2873 |
3526 |
3841 |
3985 |
4049 |
4077 |
4089 |
4094 |
4096 |
4096 |
4096 |
4096 |
4096 |
14 |
611 |
3137 |
5537 |
6931 |
7618 |
7937 |
8081 |
8145 |
8173 |
8185 |
8190 |
8192 |
8192 |
8192 |
8192 |
15 |
988 |
5769 |
10672 |
13625 |
15110 |
15809 |
16129 |
16273 |
16337 |
16365 |
16377 |
16382 |
16384 |
16384 |
16384 |
16 |
1598 |
10610 |
20570 |
26785 |
29971 |
31490 |
32193 |
32513 |
32657 |
32721 |
32749 |
32761 |
32766 |
32768 |
32768 |
By default, pyDarwin will try to search omega structure for each search block/band individually. This is only possible if all search blocks are placed in the template. If any search block is found in the tokens, the omega search will be performed uniformly, i.e. all search blocks will have the same pattern.
Individual omega search will further increase the search space size. It can be turned off by setting individual_omega_search to false.
Omega Block Search¶
Omega block search is only applicable to NLME models. It takes a diagonal Omega matrix and searches for a block Omega matrix.
To enable block search for NLME models,
Set search_omega_blocks to
truein the options file.Add one or more
#search_block(randomEffectList)to the template file and/or the tokens file, whererandomEffectListdenotes the list of random effects that one wants to search whether their associated covariance matrix (Omega) is diagonal or block.
Note
Only names, commas, and spaces (including tabs and new lines) are allowed inside the
#search_block; no comments, no nested braces.Only those random effects having diagonal Omegas are allowed inside the
search_block; if you put some random effects havingblock/same/fixedOmega, pyDarwin will halt the search.If there are random effects dependent (
same) on the ones in the#search_block, pyDarwin will halt the search.Random effects present in
#search_blockbut absent in the model (i.e., not present in anyranefstatement) will be ignored.
When creating individual models, pyDarwin puts the new ranef statement below every #search_block and fills it with corresponding Omegas, and then removes the associated random effects from the original ranef statements (basically moves the Omegas from original ranef statement to the new one). Empty diag and ranef statements are removed from the model. For example, if a template contains
ranef(diag(nV, nCl) = c(1, 1))
and the following statement is added to it
#search_block(nV, nCl))
with search_omega_blocks set to true in the options file, then pyDarwin will create two models with one having
ranef(diag(nV, nCl) = c(1, 1))
and the other one having
ranef(block(nV, nCl) = c(1, 0, 1))
Omega Band Search¶
Omega band search is only applicable to NONMEM models. It takes a diagonal OMEGA matrix and searches for band OMEGA matrices.
Band Omegas will be searched if:
The text “; search band” appears on the $OMEGA record in the template file.
The following fields have been included in the options file:
Where N is a positive integer.
Warning
The $OMEGA must appear on a separate line, e.g., $OMEGA 0.1 0.1 0.1 ; search band is not permitted.
For example, if the Omega matrix specified in the template file is:
$OMEGA ; search band
0.1
0.1
0.1
0.1
With “search_omega_bands”:true and “max_omega_band_width”: 3 specified in the options file, the search space will consist of candidate models with the following Omega structures:
Width = 0
$OMEGA BLOCK(4)
0.1
0 0.1
0 0 0.1
0 0 0 0.1
Width = 1
$OMEGA BLOCK(4)
0.1
p 0.1
0 p 0.1
0 0 p 0.1
Width = 2
$OMEGA BLOCK(4)
0.1
p 0.1
p p 0.1
0 p p 0.1
Width = 3
$OMEGA BLOCK(4)
0.1
p 0.1
p p 0.1
p p p 0.1
The actual value used for the off diagonal elements (p in example) are randomly chosen from a uniform distributions between -p and +p where p is a value less than the maximum value that results in a positive definite matrix. p will have a minimum absolute value of 0.000001 to ensure it does not have a value of 0. Each value of p in the matrix will be different.
Note
Use random_seed to ensure values of off diagonal elements are the same across subsequent searches.
Warning
If the user defines the OMEGA structure (e.g., DIAG, BLOCK, SAME) with ;; search band included, this will override the search option and the block will be used as specified in the template file, without any OMEGA band search. The OMEGA block is assumed to be variance/covariance, CORRELATON and CHOLESKY are not supported.
Note that each OMEGA in the template file can be set to searched or not searched, for example:
$OMEGA ;; search band
0.6 ; ETA(1) K23
0.5 ; ETA(2) K32
$OMEGA
0.4 ; ETA(3) CLEARANCE
0.3 ; ETA(4) VOLUME
0.2 ; ETA(5) KA
In this case the first $OMEGA block will be searched and the second will not.
Note
Comments can appear in the $OMEGA block but each row of the matrix must be on its own line.
Warning
Do not combine multiple OMEGA blocks in the template if some are BLOCK|DIAG|SAME|FIX and others are to be searched.
Omega Submatrices Search¶
OMEGA submatrices permit a wider range of OMEGA structure, and, importantly, the option to estimate fewer off diagonal elements of OMEGA. In addition to the options specified above for Omega block search (for NLME models) or Omega band search (for NONMEM models), 2 additional options should be included in the options file:
Where N is the maximum size of an OMEGA submatrix, then submatrices will be searched. OMEGA submatrices are intended to be used with OMEGA block search (for NLME models) or Omega band search (for NONMEM models) to further expand the options for OMEGA structure. Specifically,
For the source OMEGA matrix of a NONMEM model:
$OMEGA ;; search band
0.1
0.1
0.1
0.1
If band matrix search is used, for an OMEGA band width of 1, the OMEGA matrix would be:
Width = 1
$OMEGA BLOCK(4)
0.1
p 0.1
0 p 0.1
0 0 p 0.1
And with the additional sub matrix search used, this search would also include:
Width = 1
$OMEGA BLOCK(2)
0.1
p 0.1
$OMEGA BLOCK(2)
0.1
p 0.1
Resulting in one fewer variance parameters to be estimated (covariance of ETA(2) and ETA(3)).
pyDarwin Outputs¶
Console output¶
After the search command is submitted, pyDarwin first verifies that the following files and executables are available:
The template file
The tokens file
The options file
nmfe??.bat - executes NONMEM
The data file(s) for the first control that is initiated
If post run R code is requested, Rscript.exe
The startup output also lists the location of:
Data dir - folder where datasets are located. It is recommended that this be an absolute path
Project working dir - folder where template, token and options files are located, this is not set by the user
Project temp dir - root folder where model file will be found, if the option is not set to remove them
Project output dir - folder where all the results files will be put, such as results.csv and Final* files
Where intermediate output will be written (e.g., u:/user/example2/output/results.csv)
Where models will be saved (e.g., u:/user/example2/working/models.json)
NMFE??.bat (Windows) or nmfe?? (Linux) file
Rscript.exe, if used
pyDarwin provides verbose output about whether individual models have executed successfully.
A typical line of output might be:
[16:22:11] Iteration = 1, Model 1, Done, fitness = 123.34, message = No important warnings
The columns in this output are:
[Time of completion] Iteration = Iteration/generation, Model Model Number, Final Status, fitness = fitness/reward, message = Messages from NMTRAN
If there are messages from NONMEM execution, these will also be written to the console, as well as if execution failed, and, if request, if R execution failed.
If the remove_temp_dir is set to false, the NONMEM control file, output file and other key files can be found in {temp_dir}/Iteration/Model Number for debugging.
File output¶
The file output from pyDarwin is generated in real time. That is, as soon as a model is finished, the results are written to the results.csv and models.json files. Similarly, messages (what appears on the console output) are written continuously to the messages.txt file.
Messages.txt¶
The messages.txt file will be found in the working directory. This file’s content is the same as the console output.
models.json¶
The models.json will contain the key output from all models that are run. This is not a very user-friendly file, as it is fairly complex json. The primary (maybe only) use for this file is if a search is interrupted, it can be restarted, and the contents of this file read in, rather than rerunning all the models. If the goal is to make simple diagnostics of the search progress, the results.csv file is likely more useful.
results.csv¶
The results.csv file contains key information about all models that are run in a more user-friendly format. This file can be used to make plots to monitor progress of the search or to identify models that had unexpected results (crashes).
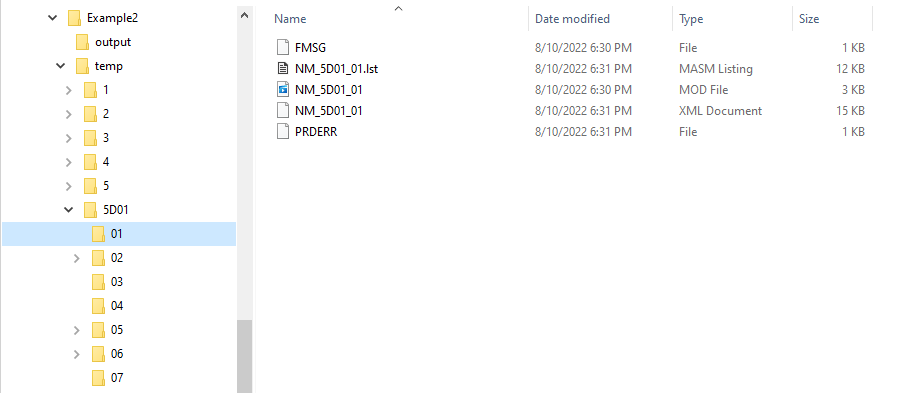
File Structure and Naming¶
NONMEM control, executable, and output file naming:
Saving NONMEM outputs¶
NONMEM generates a great deal of file output. For a search of perhaps up to 10,000 models, this can become an issue for disc space. By default, key NONMEM output files are retained. Most temporary files (e.g., FDATA, FCON) and the temp_dir are always removed to save disc space. In addition, the data file(s) are not copied to the run directory, but all models use the same copy of the data file(s).
Model/folder naming¶
A model stem is generated from the current generation/iteration and model number of the form NM_generation_model_num. For example, if this is iteration 2, model 3, the model stem would be NM_2_3. For the 1 bit downhill, the model stem is NM_generationDdownhillstep_modelnum, and for the 2 bit local search the model stem is NM_generationSdownhillstepSearchStep_modelnum. Final downhill model stem is NM_FNDDownhillStep_ModelNum. This model stem is then used to name the .exe file, the .mod file, the .lst file, etc. This results in unique names for all models in the search. Models are also frequently duplicated. Duplicated files are not rerun, and so those will not appear in the file structure.
Run folders are similarly named for the generation/iteration and model number. Below is a folder tree for Example 2.