Glossary¶
Crash Value: Anyone with experience in Population PK modeling has experience with crashes. There are many possible sources for a crash, including:
syntax errors in the control file
math errors (divide by zero, log(negative value))
numerical error in execution
Significant effort was made to capture these errors and display them to the user the console and in the messages.txt file (in the working directory).
For debugging purposes, these errors can be reproduced by going to the run directory (run_dir/generation/model_num) and rerunning the model from the command line.
In addition, errors may occur in running post run R or python code. Finally, the NONMEM or R code may time out, producing no final results. In all of these cases, the fitness/reward value assigned is called the crash_value. The crash value is set in the options file and should be substantially higher than any anticipated fitness/reward from a model that ran to completion, regardless of the “goodness” of the model.
Data Directory: Folder where datasets are located. Used if the template file specifies datasets as {data_dir}/datafileName, which is the preferred method. May be an absolute or a relative path, default value is {project_dir}. Keep in mind that NONMEM doesn’t allow dataset path to be longer than 80 characters and must be in quotes if it contains spaces.
DEAP: Distributed Evolutionary Algorithms in Python DEAP github. DEAP is a python package that includes several evolutionary algorithms, including the Genetic Algorithm - the GA option in the options file.
fitness: A number representing the overall “goodness” of the candidate. Called fitness in GA. In other algorithms, it is called “reward” or “cost”, or sometimes “loss function”. All algorithms in pyDarwin are designed to minimize this value.
full binary: The full binary is a representation of a specific model coded such that all possible values of the bit string are permitted. In general, this will result in redundancy of the matching of bit strings with the integer representation. For example, if a given token group included 3 token sets, two bits would be required (one bit can only specify two options). Two bits have 4 possible values, while only 3 are needed. Therefore, some duplication of the matching of the full binary [(0,0),(0,1),(1,0) and (1,1)] to the integer representation [1,2,3] is required. Managing this redundancy is handled internally by pyDarwin. The full binary is used only by Genetic algorithm.
The minimal binary contrasts with the full binary.
GA – Genetic Algorithm: An unsupervised search algorithm that mimics the mathematics of ‘survival of the fittest’. A population of candidates is generated randomly, and the “fitness” of each candidate is evaluated. A subsequent population of candidates is then generated using the present generation as “parents”, with selection a function of fitness (i.e., the more fit individual being more likely to be selected as parents). The parents are then paired off, undergo crossover and mutation and a new generation created. This process is continued until no further improvement is seen. In pyDarwin, GA is implemented using the DEAP package. GA on Wikipedia
GBRT – Gradient Boosted Random Tree: Similar to Random Forests , Gradient Boosted Random Trees use Gradient Boosting for optimization and may increase the precision of the tree building by progressively building the tree, and calculating a gradient of the reward/fitness with respect to each decision.
GP – Gaussian Process (Bayesian optimization): Gaussian Process is implemented in the scikit-optimize package and described here. GP is well-suited to the problem of model selection, as (according to Wikipedia) it is well-suited to black box function with expensive reward calculations. Indeed, experience to date suggests that GP, along with GA, are the most robust and efficient of the ML algorithms, especially if used in combination with a local 1 and 2 bit exhaustive search.
Home: See pyDarwin home.
Integer representation: The integer representation of a given model is what is used to construct the control file. Specifically, the integer representation is a string
of integers, with each integer specifying which token set is to be substituted into the template. For example, an integer string of [0,1,2] would substitute the
0th token set in the template for the first token group, the 1st token set for the 2nd token group and the 3rd token set for the 3rd token group. The integer representation
is managed internally by pyDarwin, and in the case of Genetic algorithm, derived from the full binary representation.
Local One bit Search: In 1 bit local search, first the minimal binary representation of the model(s) to be searched are generated. Then each bit in that bit string is ‘flipped’. So, a search with 30 bits will generate 30 models in each iteration of the 1 bit search. This process is continued, searching on the best model from the previous step until improvement no longer occurs.
For MOGA and MOGA3 the flip is performed for every objective, that gives objectives times more models in every iteration. Only unique models (not seen before) are added to the iteration to not oversaturate the algorithm. Iterations continue until the front is unchanged.
Local Two bit Search: The 2 bit local search is like the 1 bit local search except that every 2 bit change of the minimal binary representation is generated in each step, and all 2 bit change combinations are generated and run. This results in a much larger number of models to search, (N^2+n)/2. This process is again repeated until no further improvement occurs.
Local Search: It has been demonstrated that all of the available algorithms are insufficiently robust at finding the final best model. To some degree, the global search algorithms serve to essentially find good initial estimates, to make finding the global minimum (and not a local minimum) more likely. To supplement the global search algorithms, 2 local search algorithms are used. These local search algorithms systematically change each bit in the minimal binary representation of the model and run that model. The user can specify whether this local search is done on some interval or generations/iterations and/or at the end of the global search. First a 1 bit local search Local One bit Search (also called downhill search) is done, then, if requested, a Local Two bit Search is done.
Minimal Binary: The minimal binary is one of three representations of a model phenotype. The minimal binary is simply a binary that has some possible values removed to avoid duplications. For example, if the search space includes a dimension for 1, 2, or 3 compartments, 2 bits will be needed to code this. With the required 2 bits, some redundancy is unavoidable. So, the mapping might be
[0,0] -> 1
[0,1] -> 2
[1,0] -> 2
[1,1] -> 3
with 2 bit strings mapped to a value of 2. In the minimal binary, the mapping is simply
[0,0] -> 1
[0,1] -> 2
[1,0] -> 3
and a bit string of [1,1] isn’t permitted. This eliminates running the same model (from different bit strings). The minimal binary representation is used for the downhill and local 2 bit search.
The minimal binary contrasts with the full binary.
MOGA – Multi-Objecive Genetic Algorithm:
MOGA3 – Custom Multi-Objecive Genetic Algorithm:
Niche Penalty: The niche penalty is computed by first calculating the “distance matrix”, the pair-wise Mikowski distance from the present model to all other models. The “crowding” quantity is then calculated as the sum of: (distance/niche_radius)**sharing_alpha for all other models in the generation for which the Mikowski distance is less than the niche radius. Finally, the penalty is calculated as: exp((crowding-1)*niche_penalty)-1. The objective of using a niche penalty is to maintain diversity of models, to avoid premature convergence of the search, by penalizing when models are too similar to other models in the current generation. A typical value for the penalty is 20 (the default).
Niche Radius: The niche radius is used to define how similar pairs of models are. This is used to select models for the Local search, as requested, and to calculate the sharing penalty for Genetic Algorithm.
Number of effects is defined in the token sets. Each token set must include an additional final token that defines the number of effects for that token set. This token must be the final token in the token set. E.g., for a token set defining the relationship between K23 and WT, with a power model having 1 effect (one additional THETA estimated), and no relationship having 0 effects, the token set would be
"K23~WT": [
["*CWTKGONE**THETA(K23~WT)",
"(0,0.1) \t; THETA(K23~WT) K23~WT",
" effects = 1"
],
["",
"",
" effects = 0"
]
],
In general, this final token need not be included in the template.txt file, but it may be included if desired, if it is commented out in the code, e.g.,
$EST METHOD=COND INTER MAX = 9999 MSFO=MSF1 PRINT = 10
$COV UNCOND PRINT=E PRECOND=1 PRECONDS=TOS MATRIX = R
$TABLE REP ID TIME DV EVID NOPRINT FILE = ORG.DAT ONEHEADER NOAPPEND
;;number of K32 {K23~WT[3]}
which would generate control file text of
$EST METHOD=COND INTER MAX = 9999 MSFO=MSF1 PRINT = 10
$COV UNCOND PRINT=E PRECOND=1 PRECONDS=TOS MATRIX = R
$TABLE REP ID TIME DV EVID NOPRINT FILE = ORG.DAT ONEHEADER NOAPPEND
;;number of K32 effects = 1
and
$EST METHOD=COND INTER MAX = 9999 MSFO=MSF1 PRINT = 10
$COV UNCOND PRINT=E PRECOND=1 PRECONDS=TOS MATRIX = R
$TABLE REP ID TIME DV EVID NOPRINT FILE = ORG.DAT ONEHEADER NOAPPEND
;;number of K32 effects = 0
respectively, for these token sets.
The number of effects can be larger than 1. For example, if an Emax relationship between parameters and covariates is to be tested, two THETAs will be added, then the number of effects may be 2, e.g.,
"K23~WT": [
["*CWTKGONE * THETA(K23~WTmax)/ CWTKGONE + THETA(K23~WT50)/",",
"(0,0.5) \t; THETA(K23~WTmax) K23~WT max effect" \n(0,1) \t; THETA(K23~WT) K23~WT50 50% effect",
" effects = 2"
],
["",
"",
" effects = 0"
]
],
Parameter sorting: The tokens are first merged into the template file. In this merged file, the parameters in the searched text are indexed only with text, e.g., THETA(ALAG). This is necessary as the integer indices assigned to each parameter cannot be determined until the control file is merged. Once this is done, the number and sequence of searched THETA/OMEGA/SIGMA values in the control file can be determined and the correct parameter indices assigned. Essential rules for parsing the merged template are:
Fixed parameter initial estimates should be placed before the searched parameter initial estimates. For example, the following is not recommended (although it may work).
$THETA
(0,1) ; THETA(1) Clearance
{ALAG[2]}
(0,1) ; THETA(2) Volume
A searched parameter initial estimate ({ALAG[2]}) occurs before a fixed initial estimated ((0,1) ; THETA(2) Volume).
Each parameter initial estimate must be on a separate line. Parameter estimates must be enclosed in parentheses, e.g., (0,1).
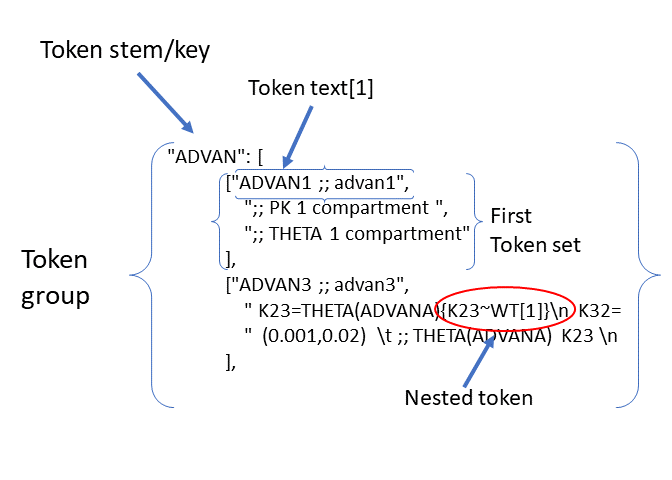
Nested Tokens: pyDarwin permits nested tokens to be used in the tokens file. This permits one token to contain another token, to an arbitrary level. Note that
using nested tokens does not reduce the search space size, it only reduces the number of token sets the user needs to generate, and simplifies the logic (although, commonly, the logic quickly
becomes impenetrable). For example, assume that the search is to contain one compartment
(ADVAN2) and two compartment (ADVAN4), and if ADVAN4 is selected, search whether K23 and K32 are functions of weight. K23 is not a parameter of a one compartment model. One option would be to simply write out
all possible models:
1 compartment
["ADVAN2 ;; advan2",
";; PK 1 compartment ",
";; THETA 1 compartment"
],
2 compartment – without K23~weight
["ADVAN4 ;; advan4",
"K23=THETA(ADVANA)\n K32=THETA(ADVANB)",
"(0.001,0.02) \t ; THETA(ADVANA) K23 \n (0.001,0.3) \t ; THETA(ADVANB) K32 "
],
2 compartment – with K23~weight
["ADVAN4 ;; advan4",
"K23=THETA(ADVANA)*CWT**THETA(K23~WT)\n K32=THETA(ADVANB)*CWT**THETA(K23~WT)",
"(0.001,0.02) \t ; THETA(ADVANA) K23 \n (0.001,0.3) \t ; THETA(ADVANB) K32 \n (0,0.1) \t; THETA(K23~WT) K23~WT" "
],
2 bits would be required to specify these 3 options.
An alternative is to have one token group for a number of compartments:
1 compartment vs 2 compartment, and have the K32~WT nested within the ADVAN4
["ADVAN2 ;; advan2",
";; PK 1 compartment ",
";; THETA 1 compartment"
],
["ADVAN4 ;; advan4",
"K23=THETA(ADVANA)**{K23~WT[1]}**\n K32=THETA(ADVANB)**{K23~WT[1]}**",
"(0.001,0.02) \t ; THETA(ADVANA) K23 \n (0.001,0.3) \t ; THETA(ADVANB) K32 \n{K23~WT[2]} \t ; init for K23~WT "
],
and another token set (nested within the ADVAN token set) for K23 and K32~WT
[
["",
""
],
["*WTKG**THETA(K23~WT)",
"(0,0.1) \t; THETA(K23~WT) K23~WT"
]
],
This also requires 2 bits, one for the ADVAN token group, one for the K23~WT token group. Using nested tokens can reduce the number of token sets in a token group, at the expense of more token groups. While more than one level of nested tokens is permitted, the logic of constructing them quickly becomes very complicated.
The full example is given in example 4.
nmfe_path: The path to nmfe??.bat (Windows) or just nmfe?? (Linux). Must be provided in the nmfe_path. Only NONMEM 7.4 and 7.5 are supported.
Options File: Specifies the options for the search, including the algorithm, the fitness/reward criteria, the population size, the number of iterations/generations and whether the downhill search is to be executed.
results.csv and Final .mod and .lst files are saved. Default value is
‘{working directory}/output’.{project_dir}/output may be used if you want to version control the project and the results.Phenotype: The integer string representation for any model.
Project Directory - Folder where the template, token, and options files are located (and maybe datasets, see Data Directory).
Can be provided as an argument for run_search_in_folder or determined by path to options.json (as parent folder). This cannot be set in options file.
pyDarwin home Default location for every project’s {working_dir}. Usually is set to <user home dir>/pydarwin. Can be relocated using PYDARWIN_HOME.
Reward: A number representing the overall “goodness” of the candidate, the sum of -2LL and the user-defined penalties. Called “fitness” in GA.
RF – Random Forest: Random Forest consists of splitting the search space (based on the “goodness” of each model, in this case), thus continuously dividing the search space into “good” and “bad” regions. As before, the initial divisions are random, but become increasingly well-informed as real values for the fitness/reward of models are included.
scikit-optimize: Optimization package The python package that includes GP, RF, and GBRT.
temp_dir: Folder where all iterations/runs are performed, i.e., where all NONMEM files are written. Default value is ‘{working_dir}/temp’. May be deleted after search finished/stopped, if remove_temp_dir is set to true.
Template: A text string, saved in the template file that forms the basis for the models to be run. The template file is similar to a NONMEM control file, but with tokens that are replaced by text strings specified in the tokens file.
Token: A token is a text string that appears in the Template. The format of the string is {token stem [index]}, where token stem identifies the token group and index identifies which token key-text pair within the token set is to be substituted.
Tokens file: See “Tokens file”.
Token group: A token group is a collection of token sets that defines a single dimension in the search space. For any model, exactly one of the tokens sets will be selected to be substituted into the template file.
Token set: One for each option in the dimension.
Token key-text pair: A token set contains two or more token key-text pairs. These pairs are very analogous to JSON key-value pairs, except that only text values are permitted. For each token key-text pair, the text {token stem [n]} in the template is replaced by the corresponding values in the token key-text pair. Note that the token key is surrounded by curly braces in the template file. For example, if the template contains these two tokens
{ALAG[1]}
in the $PK block
and
{ALAG[2]}
in the $THETA block, the token stem would be ALAG. Again, note that in the template file the “token stem[n]” is enclosed in curly braces. N is the index of the token within the token set. While indices to token can be duplicated and indices can be skipped, it is recommended that they start at 1 and be numbered sequentially through the template file. The ALAG token group would be required in the tokens files. Exactly one token set would be selected (by the search algorithm) for substitution into the template file. If the first token set is selected, and this token set contains the following token key-text pairs
ALAG[1] -> "ALAG1=THETA(ALAG)"
ALAG[2] -> "(0,1) ;; initial estimate for ALAG1"
the text “ALAG[1]” in the template file would be replaced by “ALAG1=THETA(ALAG)” and
the “ALAG[2]” text in the template would be replaced by “(0,1) ;; initial estimate for ALAG1”. This would
result in syntactically correct NMTRAN code (except that the index to THETA is still a text string). The appropriate
index for THETA can be determined only after all the features/token sets are selected. This is handled by pyDarwin. Similar
logic (ETAs index by text strings, which are replaced by integers) applies for ETAs and EPSs. It is most convenient to use the token stem to
index the parameters, e.g., for the CL~WT tokens set, one might use THETA(CL~WT). If more than one THETA is used in a token set, one can
simply add an integer (e.g., THETA(CL~WT1) and THETA(CL~WT2)), but the THETA text indices must be unique, to generate unique integer values. Any
duplication of THETA text indices is permitted (e.g., if you want the same exponent for CL and Q) but will result in duplication of the integer indices, e.g.,
{*WTKG**THETA(CL~WT)} ;; for clearance
and
{*WTKG**THETA(CL~WT)} ;; for Q
would result in
CL=THETA(1)*WT**THETA(2) ;; for clearance
and
Q =THETA(2)*WT**THETA(2) ;; for Q
Duplicate text indices will yield duplicate integer indices. By the same logic, comments can be put into initial estimates by including THETA(CL~WT) after a “;” in the $THETA block, e.g.,
(0,0.75) \t; THETA(CL~WT) exponent on clearances
will result in
(0,0.75) ;THETA(2) exponent on clearances
as the THETA(CL~WT) is similarly replaced by THETA(2).
Token stem: The token stem is a unique identifier for the token group. This text string is used to link the tokens in the template file to the token sets. In the json code file for tokens:
Tournament Selection An algorithm used in GA when there are two or more “parents”. The one with the highest fitness (lowest penalized -2LL) wins and enters into the next generation.
Working directory Folder where all project’s intermediate files are created, such as models.json (model run cache), messages.txt (log file), interim model files, and stop files. It is also a default location of output and temp folders. Can be set with working_dir option. By default, it is set to ‘<pyDarwin home>/{project_stem}’.