Plot parameter estimates against covariates
nlme.par.vs.cov.RdUse to create a stack of plots of parameter estimates plotted against covariates.
Arguments
- xpdb
An xpose database object.
- covColNames
Character vector of covariates to build the matrix.
- nrow
Number of rows.
- ncol
Number of columns; if ncol=1, each gtable object is treated separately.
- ...
Parameters to be passed to
ggarrange().
Value
List of gtable
Examples
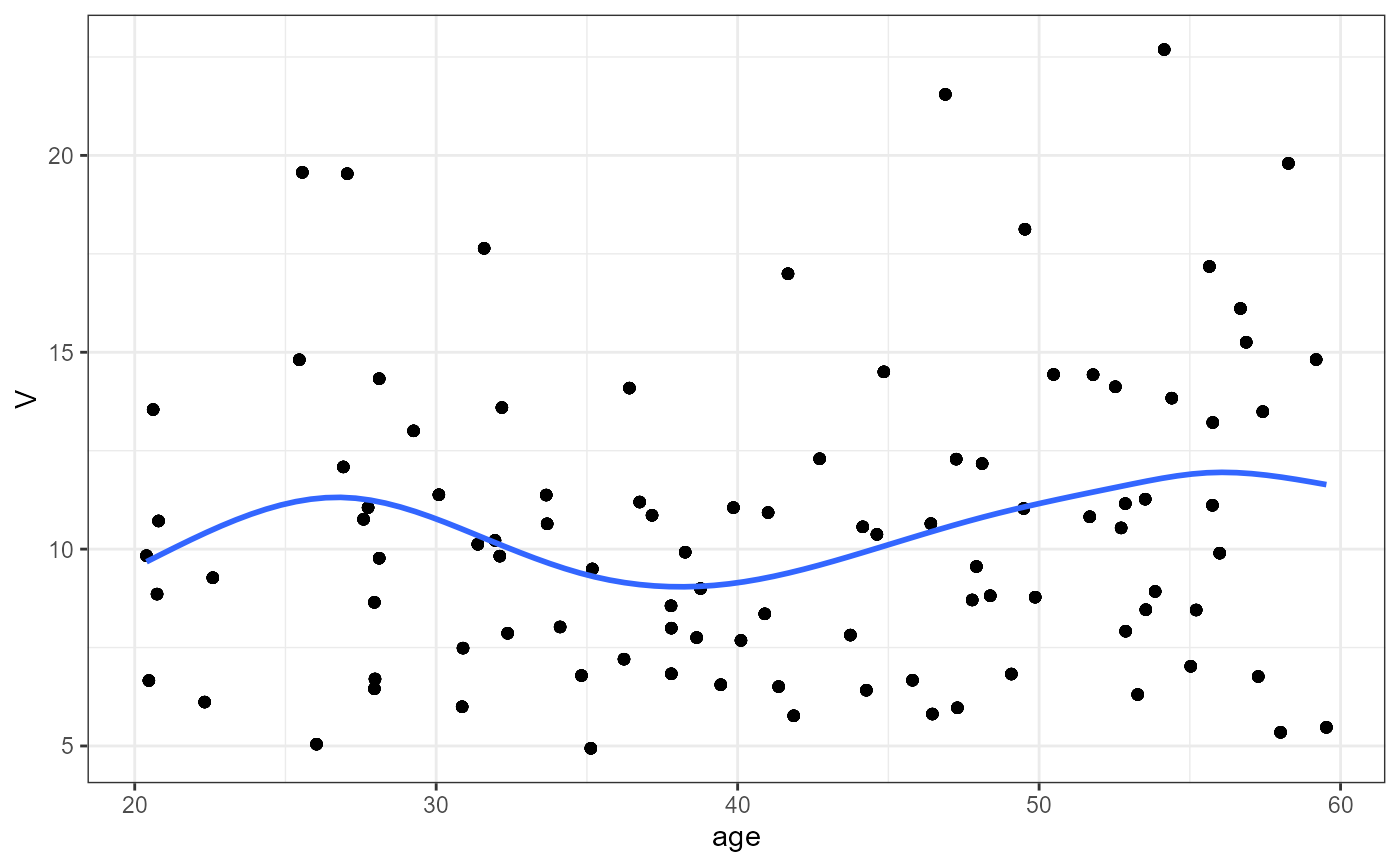
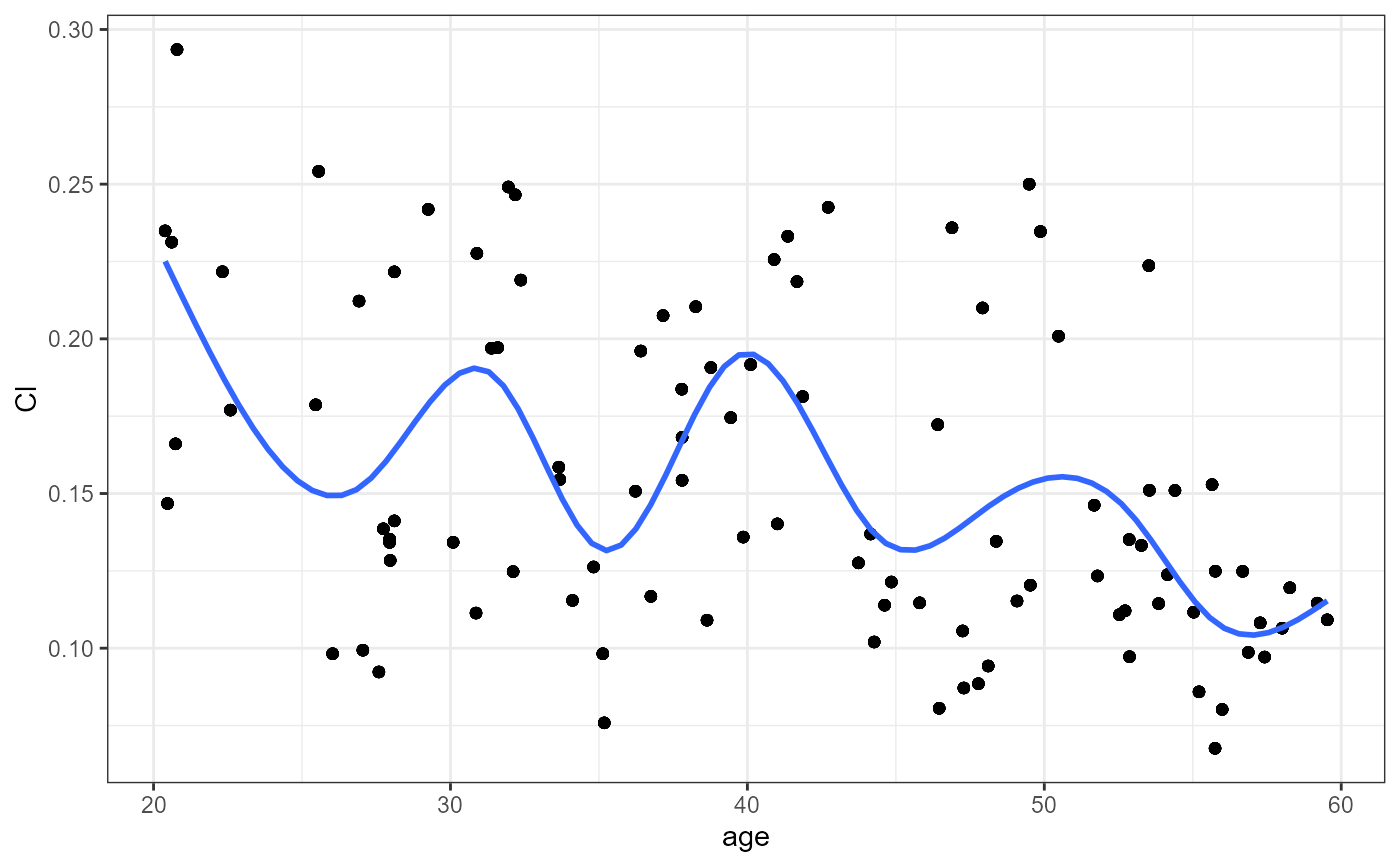
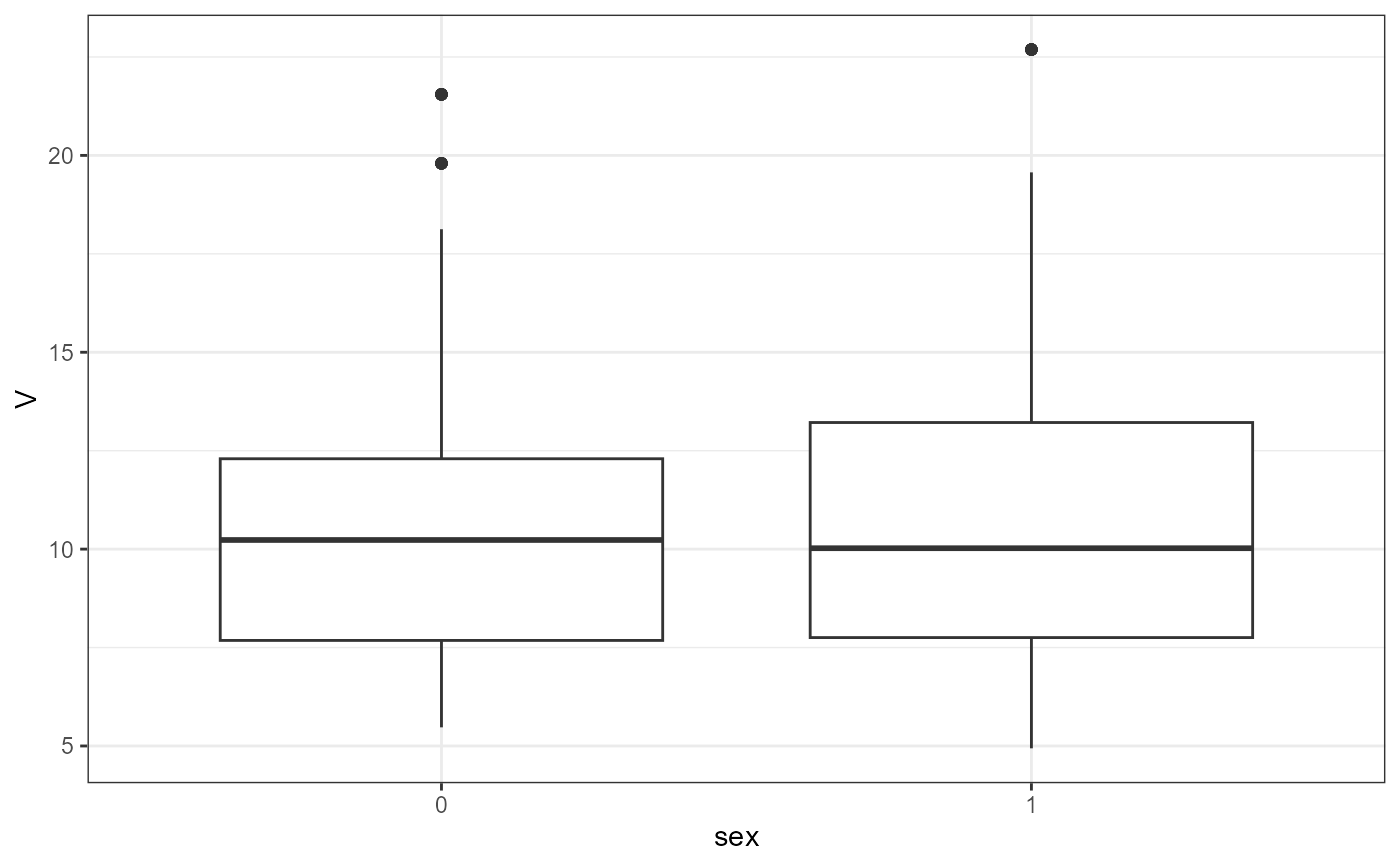
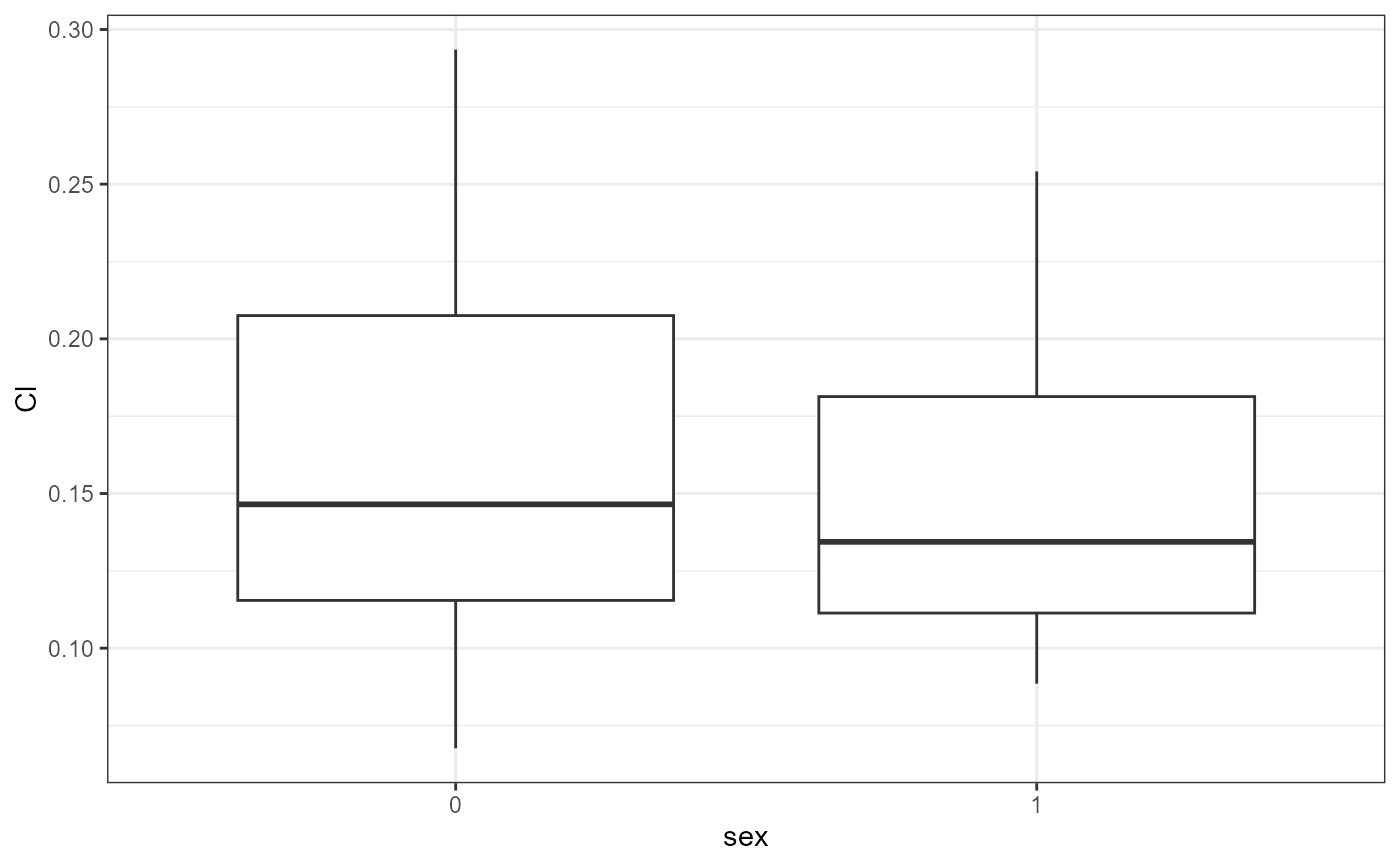
nlme.par.vs.cov(
xpdb = xpdb_ex_Nlme,
covColNames = c("sex", "wt", "age")
)
#> `geom_smooth()` using method = 'gam' and formula = 'y ~ s(x, bs = "cs")'
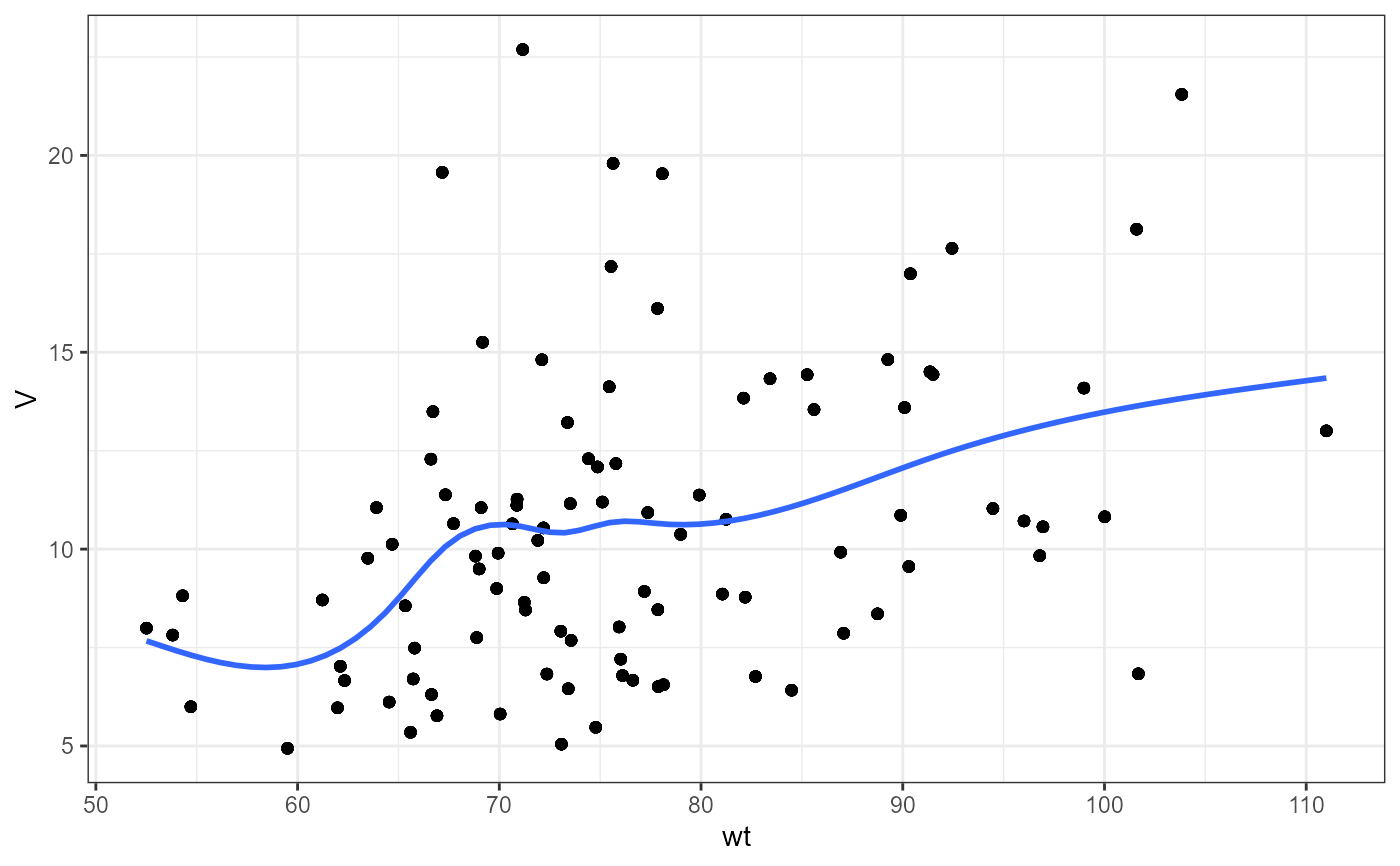 #> `geom_smooth()` using method = 'gam' and formula = 'y ~ s(x, bs = "cs")'
#> `geom_smooth()` using method = 'gam' and formula = 'y ~ s(x, bs = "cs")'
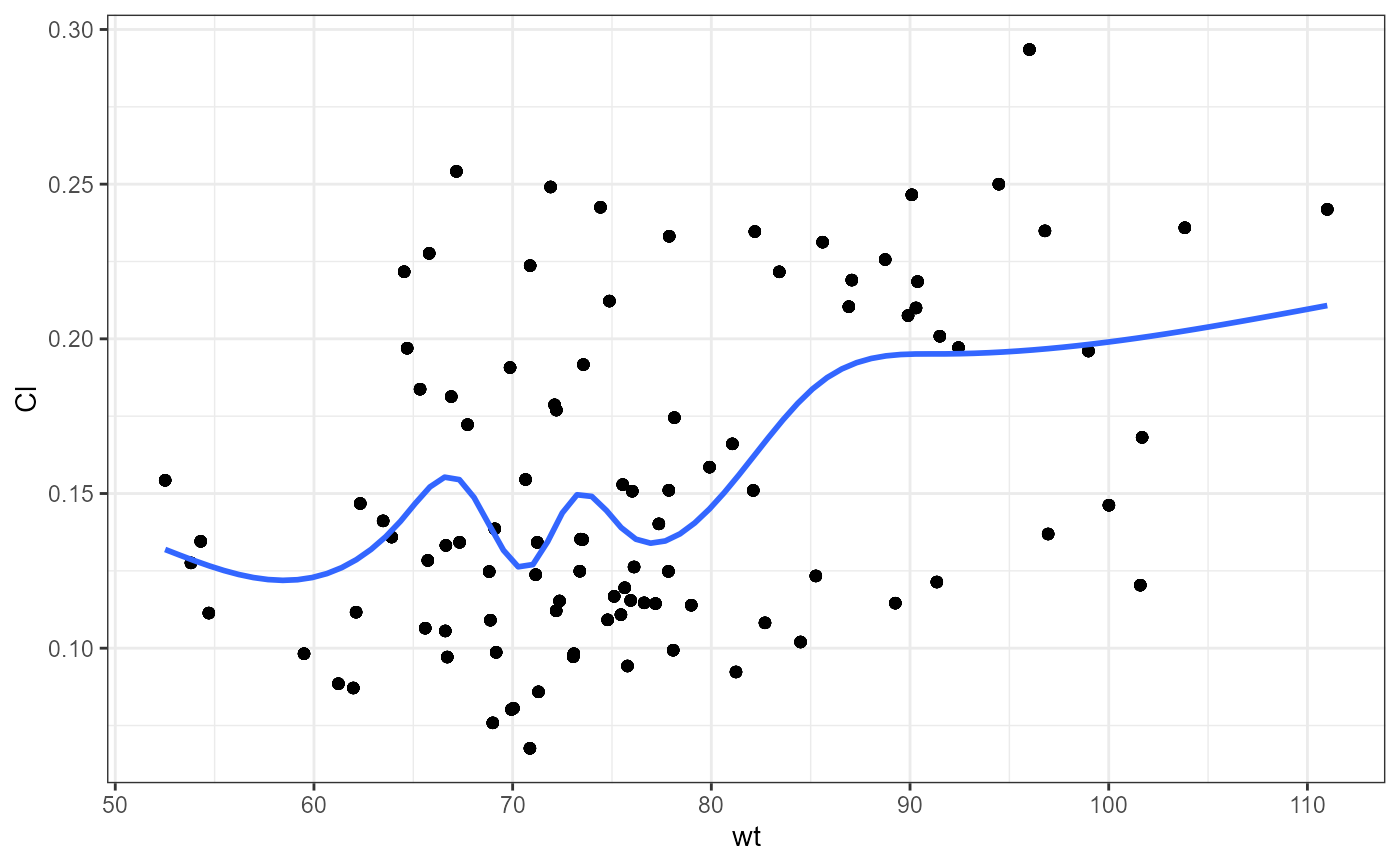 #> `geom_smooth()` using method = 'gam' and formula = 'y ~ s(x, bs = "cs")'
#> `geom_smooth()` using method = 'gam' and formula = 'y ~ s(x, bs = "cs")'
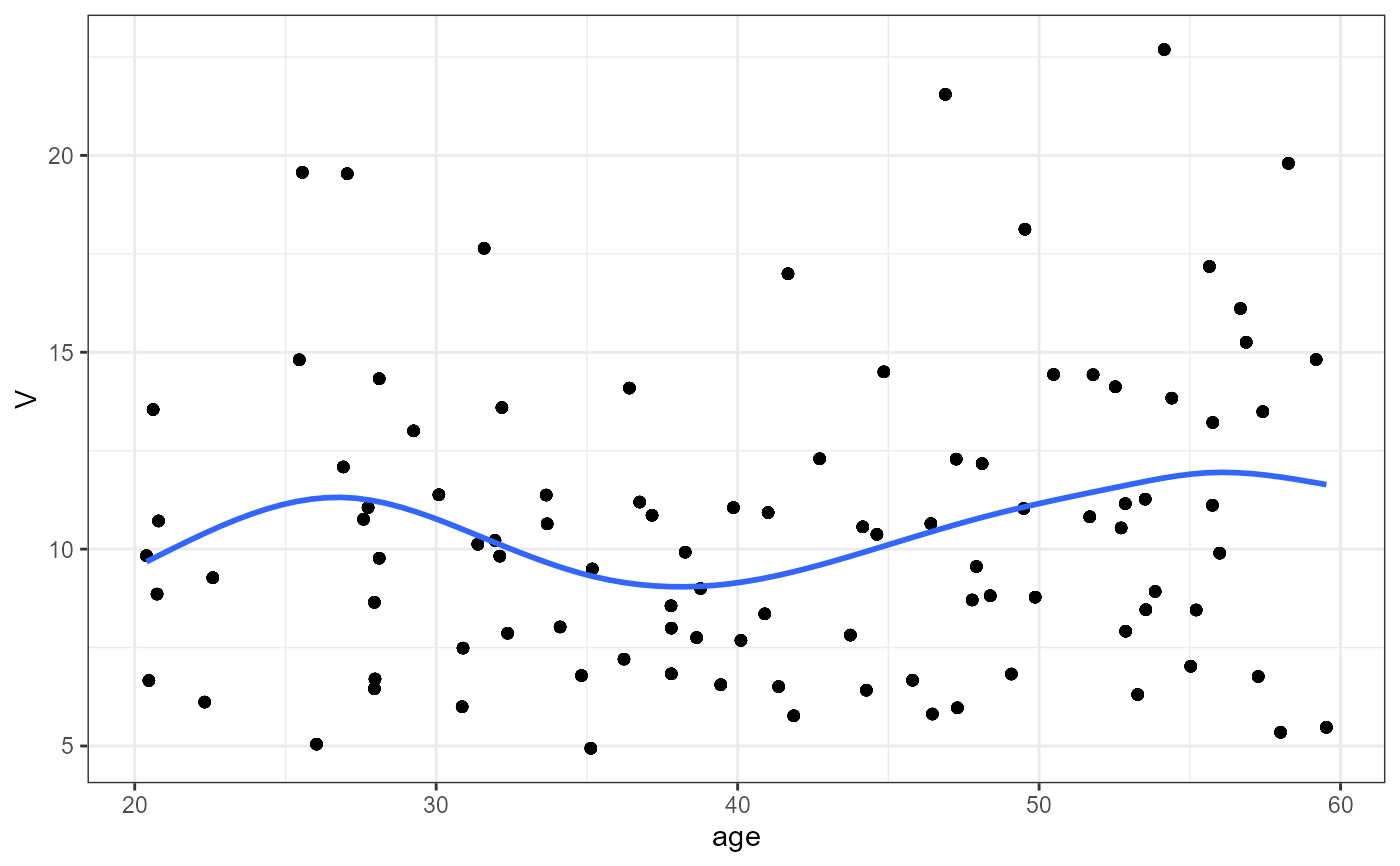 #> `geom_smooth()` using method = 'gam' and formula = 'y ~ s(x, bs = "cs")'
#> `geom_smooth()` using method = 'gam' and formula = 'y ~ s(x, bs = "cs")'
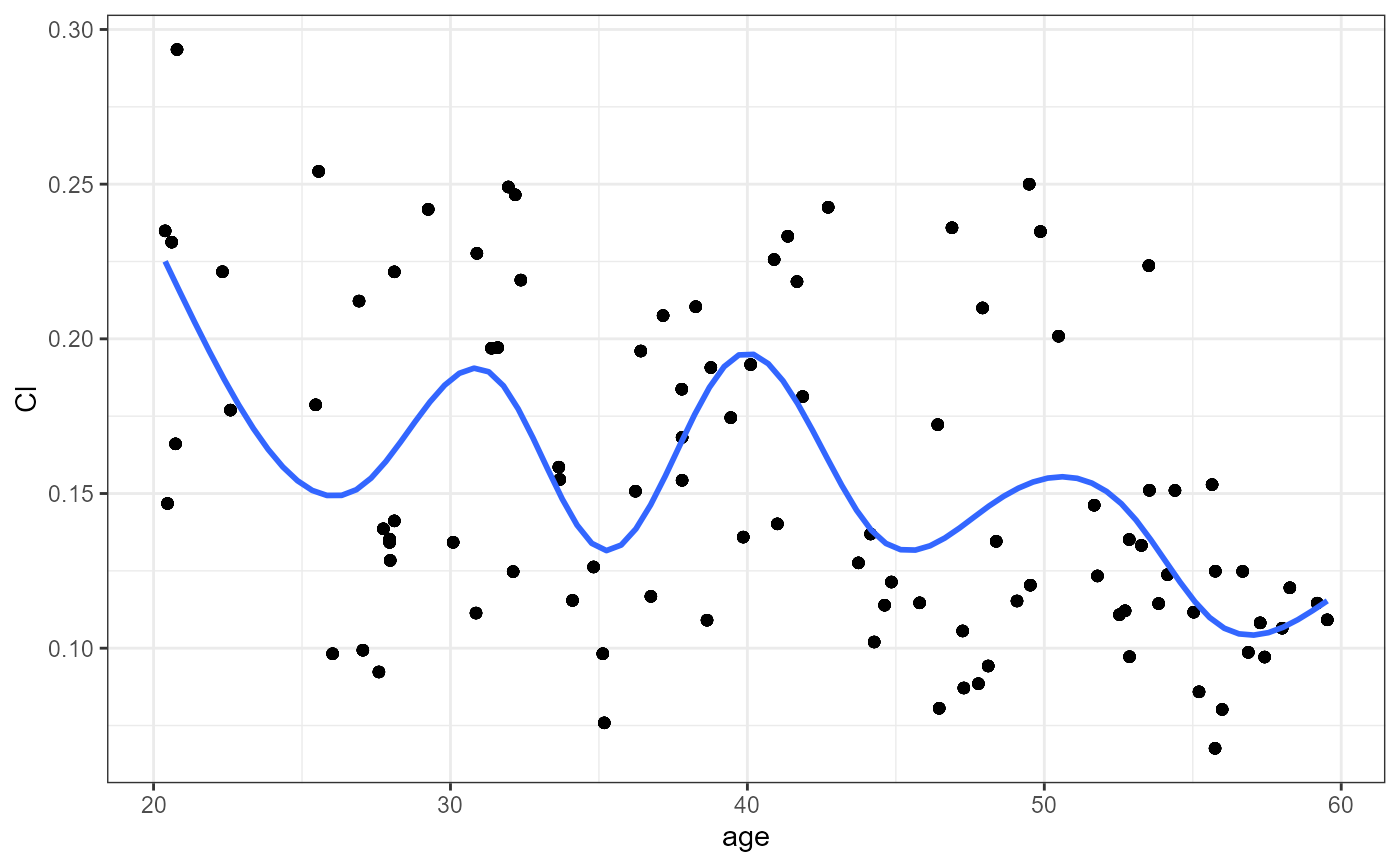
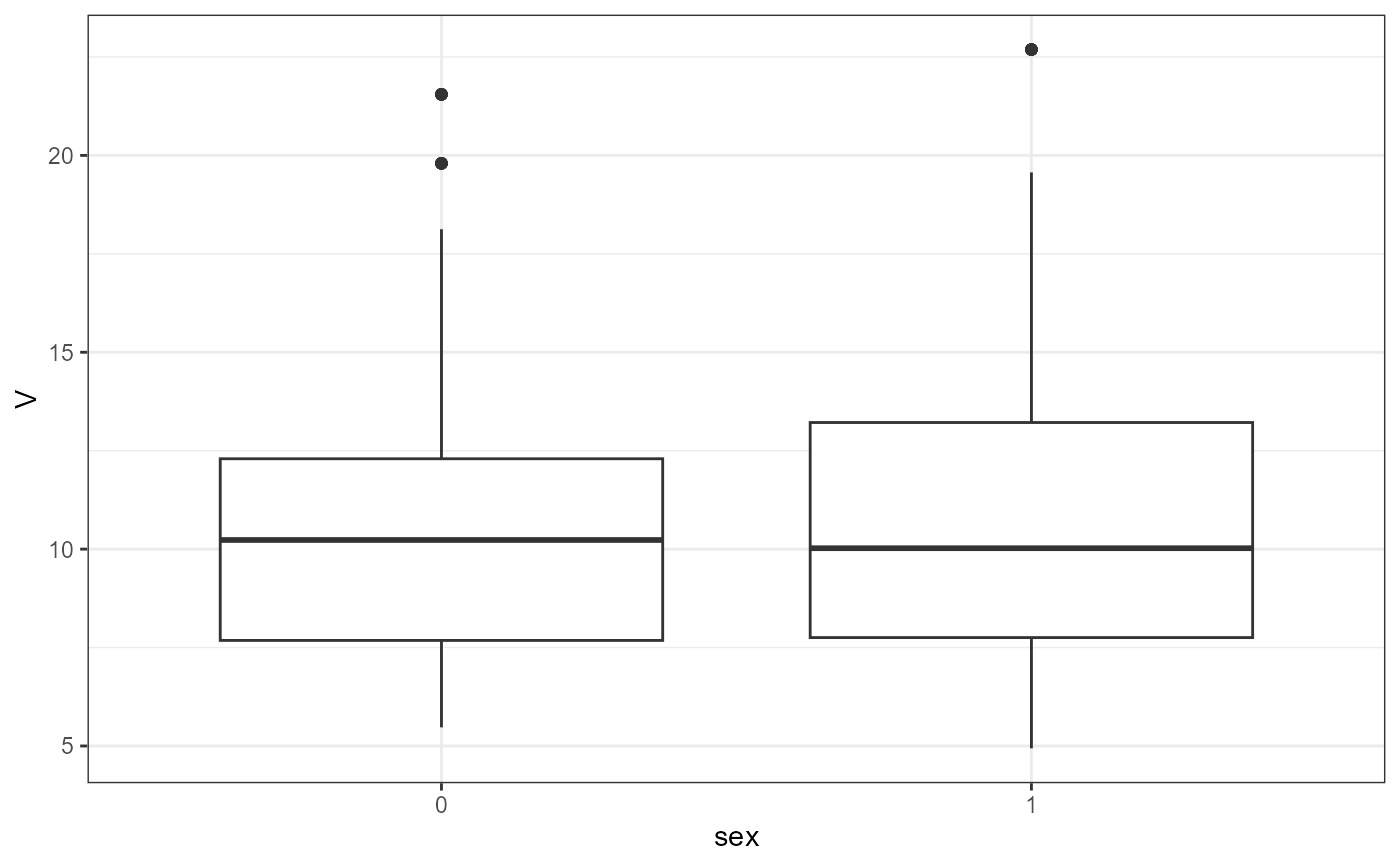
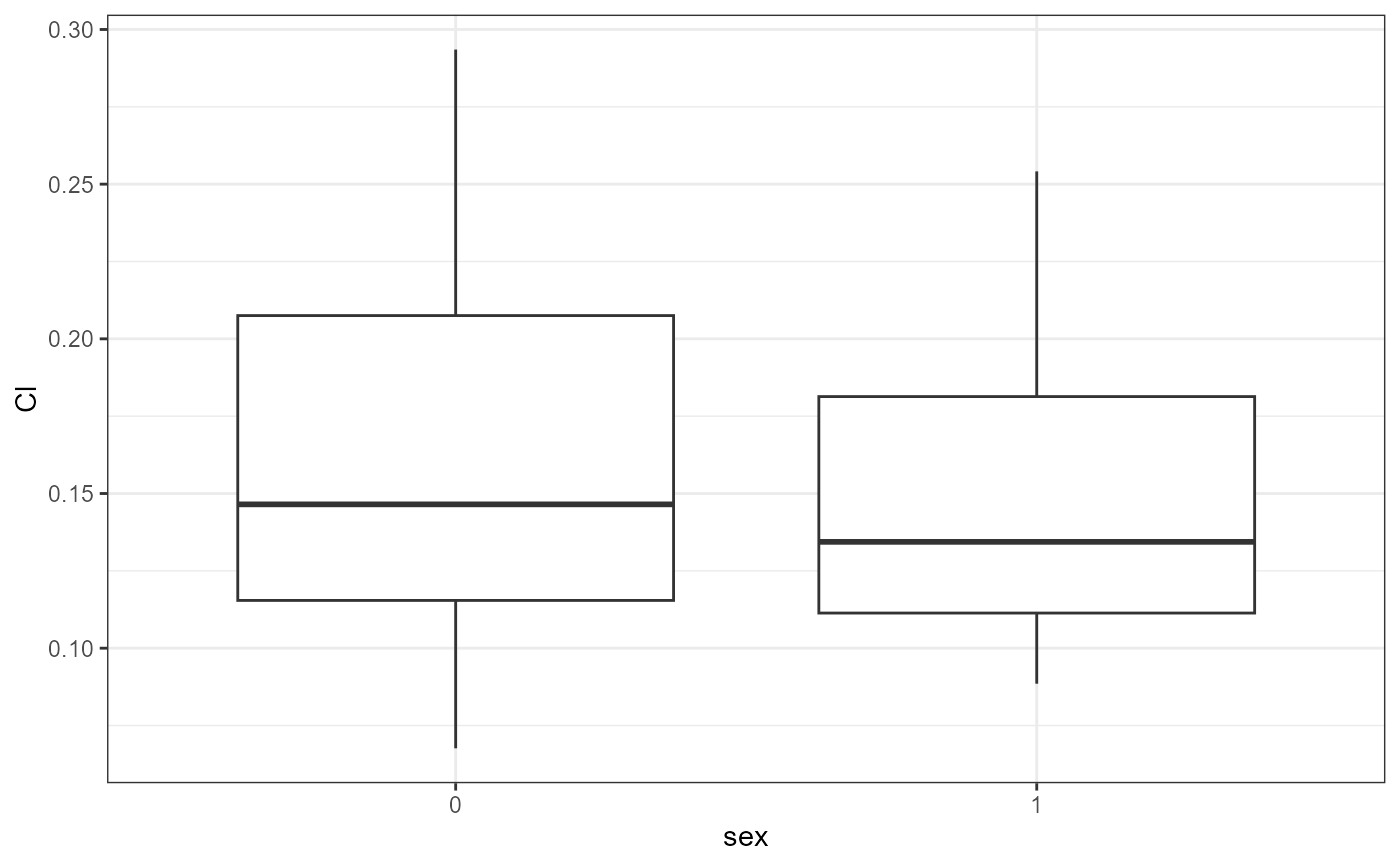 #> [[1]]
#> [[1]]
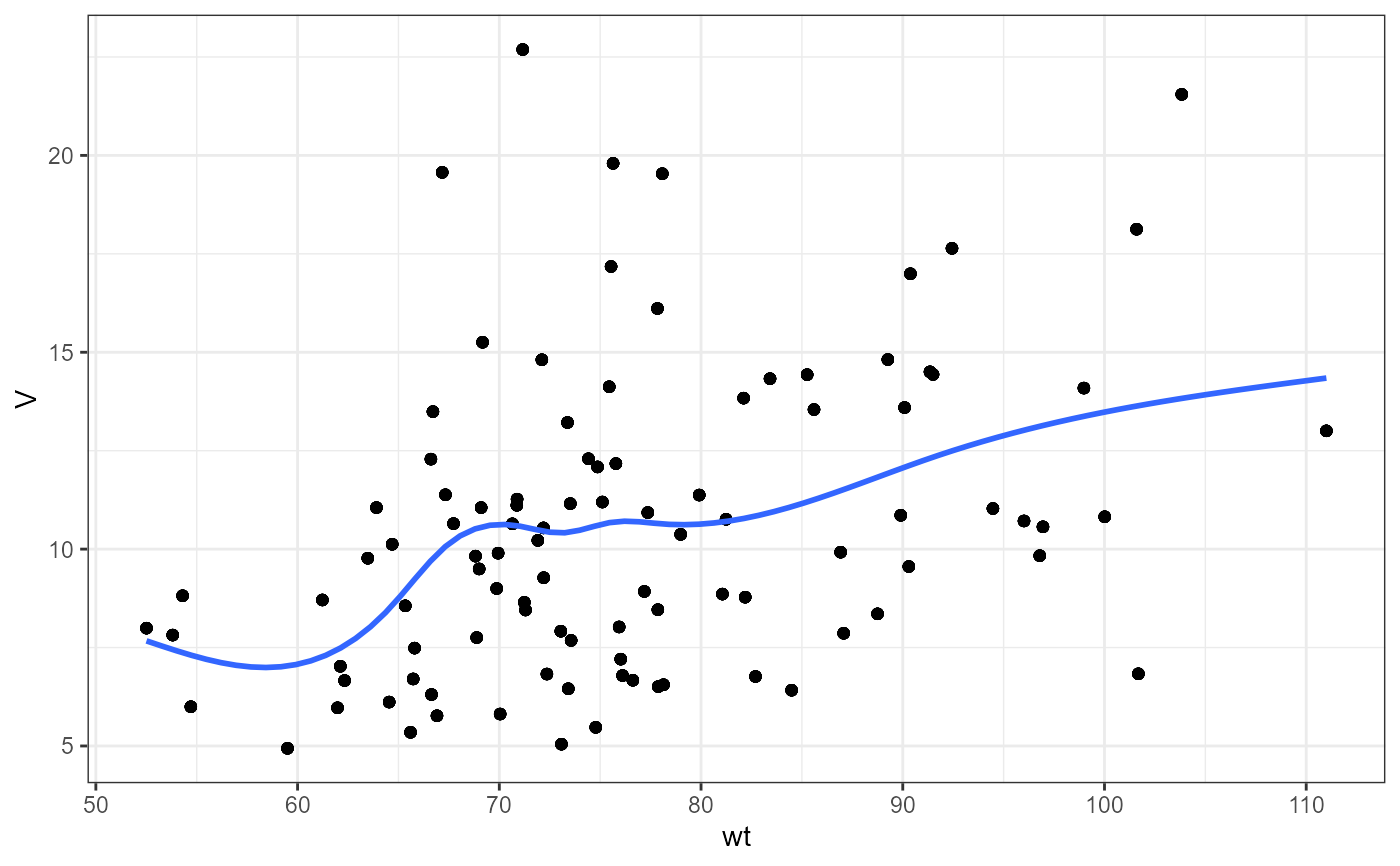 #>
#> [[2]]
#>
#> [[2]]
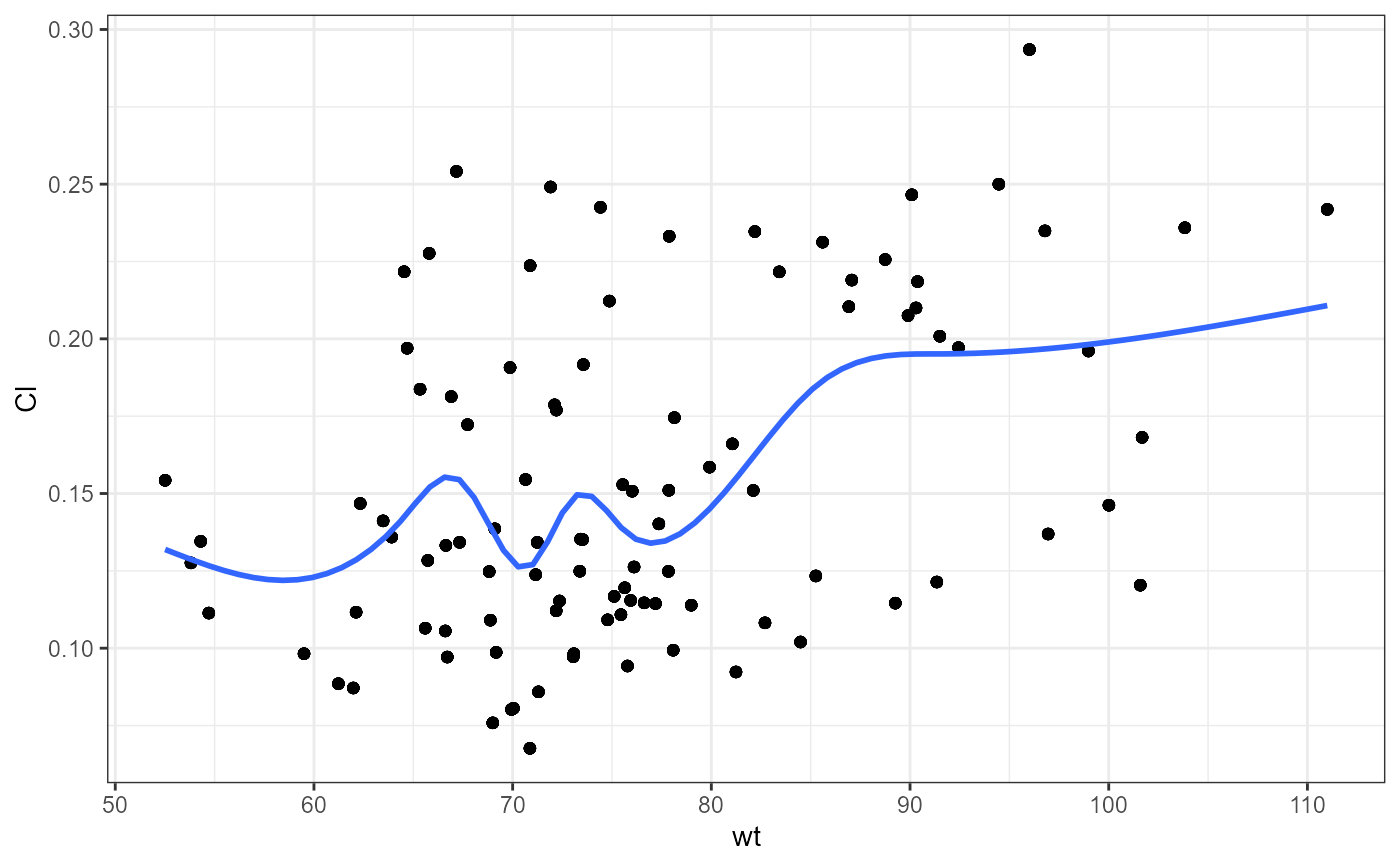 #>
#> [[3]]
#>
#> [[3]]
 #>
#> [[4]]
#>
#> [[4]]
 #>
#> [[5]]
#>
#> [[5]]
 #>
#> [[6]]
#>
#> [[6]]
 #>
#>