Integral for Validated R Package Repositories
Source:vignettes/integral_package_repositories.Rmd
integral_package_repositories.RmdOverview
Among its many capabilities, Integral can function as a centralized repository for storing, distributing, and version-controlling validated R packages, which is critical for ensuring reproducibility in analyses, especially in light of potential changes to package versions over time. By leveraging the renv and miniCRAN packages, users can create a “snapshot” of the R packages used in an analysis and seamlessly upload them to Integral from R. Importantly, when the analysis needs to be reproduced at a later time, the package repository can be downloaded from Integral, allowing other users to easily install the required packages and recreate the exact R environment used by the original analyst. Hosting these package snapshots within Integral provides a safeguard against the challenges posed by updates or removals of packages from CRAN, enabling consistent and reproducible workflows across teams and projects.
In this vignette, we demonstrate a minimally reproducible example of how to create a portable package repository using miniCRAN and save it to Integral. This approach simplifies the process of sharing and reproducing R environments, ensuring a seamless experience for all users.
Setup
To create a snapshot of the environment, the renv and miniCRAN packages will need to be installed locally.
install.packages(c("renv", "miniCRAN"))While not required, the renv package works best when
used in an R project environment. An R project creates a
self-contained working directory for your analysis, which includes a
.Rproj file to store project-specific settings and
metadata. If you are not performing your analysis within a designated R
project, make sure that your current working directory is not cluttered
with irrelevant files. renv will check all package
dependencies in the defined project or environment, so it is imperative
that only relevant packages exist within this working directory.
For the purpose of this vignette, we will set our working directory
to be a folder that contains a single file called
dependencies.R, which will simulate an analysis script.
setwd("~/original_analysis")
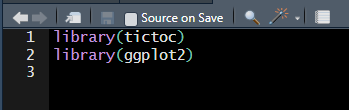
This file consists of two lines of code that are calling packages, similar to how one would document any packages needed at the top of their analysis script.
Snapshotting Current Packages
Now that we are working in a folder designed specifically for our
analysis, we are ready to take a snapshot of the packages we need. Using
the snapshot() function from the renv
package, we can create a JSON lockfile, which contains
information on all the packages that are being used in our current
folder. In addition to the packages we explicitly named, it also records
any of our packages’ required dependencies.
renv::snapshot()The snapshot() function accepts a type
argument, which dictates how the function decides which packages to
include in the lockfile. Values passed to this argument can be either
“implicit” or “explicit”.
Implicit Packages
The default behavior is the same as specifying
snapshot(type = "implicit"), which will record all the
packages returned by renv::dependencies(). It will crawl
through all the R scripts in your project, looking for any calls to
packages with library() as well as package
NAMESPACE prefixes (e.g.,
package::function()). This behavior works well for a single
analysis or project, where a user wants to create a repository
containing only the packages required to re-run the analysis.
Explicit Packages
Alternatively, you can use snapshot(type = "explicit")
to create the lockfile. This behavior only checks packages recorded in
the DESCRIPTION file. While the DESCRIPTION
file is most often used in packages, it can also be added to an R
project (usethis::use_description(check_name = FALSE)).
snapshot() will check the Imports section of
the DESCRIPTION file to determine what packages to capture.
This method is more applicable for an admin who wants to validate and
capture an entire suite of R packages to be used in a variety of use
cases across multiple users.
Creating a Local Package Repository
Now that we know what packages are needed, we will create a local repository of these packages using the miniCRAN package. The functions from this package require a repository or CRAN mirror (typically your regional CRAN repository) to be defined as an argument.
CRAN <- c(CRAN = "https://cloud.r-project.org")We can now parse the lockfile using the lockfile_read()
function, and then extract the names of the packages being used. As an
additional safeguard, we will also check for dependencies using the
pkgDep() function from miniCRAN.
# Parse lockfile
lockfile <- renv::lockfile_read()
# Parse package names
pkgs <- names(lockfile$Packages)
# Check for package dependencies, while ignoring suggested packages
pkgList <- miniCRAN::pkgDep(pkgs, repos = CRAN, type = "source", suggests = FALSE)Since this repository will always be available on Integral, there is
no need to keep in a permanent location locally; we will create a
temporary directory called miniCRAN to serve as our repository
to export. With our temporary directory created, we will use the
makeRepo() function from miniCRAN to
download all the packages in our list to our temporary repository.
# Create temporary directory
miniCRAN_pth <- file.path(tempdir(), "miniCRAN")
dir.create(miniCRAN_pth)
# Download packages and create the repository
miniCRAN::makeRepo(pkgList, path = miniCRAN_pth, repos = CRAN, type = c("source", "win.binary"))An important detail to note is that makeRepo() installs
the most up to date version of packages. As binaries for old packages
are generally not available from CRAN, adding old packages will require
additional manual work, so using the most up to date versions of your
chosen packages will allow for the easiest workflow. For ways to use
previous package versions or to fine tune your repository, see the Advanced
Techniques for Repository Management vignette.
Uploading the Package Repository to Integral
With a local folder containing all the packages needed to recreate
our analysis, we can now zip our folder and upload it to Integral. This
will allow all future Integral users to download the repository and use
the same packages used in the original analysis. We should also upload
our renv.lock file as well, as it contains valuable
information about our packages and R environment.
library(Certara.IntegralR)
# Change to the miniCRAN directory temporarily. This makes handling paths in zip() easier.
old_wd <- setwd(miniCRAN_pth)
# Zip the miniCRAN directory.
# List all the files (recursively) in the new working directory,
files_to_zip <- list.files(".", recursive = TRUE, full.names = FALSE)
# Create the path name of the zipped repo
zipped_repo <- file.path(tempdir(), "project_001_miniCRAN.zip")
# Zip the listed files to the defined folder path
utils::zip(
zipfile = zipped_repo,
files = files_to_zip
)
# Change back to our original working directory
setwd(old_wd)
# Upload the zipped repo along with the renv.lock file
integral_upload(
file = c(zipped_repo, "renv.lock"),
path = "My Study/R Package Repositories/Project 001",
reason = "Create an R package repository to recreate analysis"
)Downloading and Using the Repository
Downstream users can now download the saved package repository and use it as their installation source for packages. This will provide them with the exact same version of packages used in the original analysis. This local repository will be stored within the project folder and all packages will be installed from here instead of from the master CRAN location.
# Move to a clean folder to recreate analysis
setwd("~/new_analysis")
# Download the repository and lockfile to the new folder
Certara.IntegralR::integral_download(
file = c("My Study/R Package Repositories/Project 001/renv.lock",
"My Study/R Package Repositories/Project 001/project_001_miniCRAN.zip"),
path = getwd()
)
# Unzip the zipped folder to a local sub folder by the same name
unzip(
zipfile = "project_001_miniCRAN.zip",
exdir = "project_001_miniCRAN"
)With the package repository downloaded locally, we will activate and
load a renv environment with the activate() and
load() functions from renv. This will create
a local folder structure that will serve as the package library that
packages from our local repository will be downloaded to. All calls to
library() a package will now pull packages in from this
local library. After doing this, we can see that our library paths have
been changed to our new local library, as well as an additional
temporary one that holds base R packages associated with our current R
version.
# Activate a new renv
renv::activate()
renv::load()
# Checking that our .libPaths() have changed to a renv library
.libPaths()Finally, we need to restore our environment to the one
described in the lockfile. While we have these packages downloaded in a
local repository, they have not actually been installed. Using
the restore() function from renv, we will
supply the lockfile that describes the packages needed, along with the
location of our local repository from which to download the
packages.
renv::restore(repos = "project_001_miniCRAN")We now have our local environment set up to recreate our original
analysis. Calling library() to our original two packages
(tictoc and ggplot2) will be successful,
while calling it for a package that was not listed in our lockfile will
return an error.
Once you are done working in this analysis folder, you can use
renv::deactivate() to reset your .libPaths()
to your normal system library. This will retain your local
renv library in the folder, allowing you to easily come
back and continue work on the project at a future date by simply using
renv::activate() again.