Advanced Techniques for Repository Management
Source:vignettes/advanced_repository_techniques.Rmd
advanced_repository_techniques.RmdOverview
While snapshotting a repository is much easier when planned in advance, it is likely that you will have projects that are already completed that need to be archived. These might contain older versions of packages that may be hard to source. Alternatively, you may want to create a package repository that only contains packages your organization has approved and validated. Using tested and validated versions of packages is crucial in creating reproducible and reliable analyses, and Integral serves as a key component in creating a reliable repository for future use. Once you have gone through the effort to create a validated set of R packages, you will want to save them to Integral to enable downstream users easy access to these reliable and tested versions. In this vignette we will cover how to build a repository with specific package versions, along with strategies to modify and update an already built local repository.
A useful tool for assessing the risks of various packages while
building your organizations repository is the riskmetric
package. More information about risk management and
riskmetric can be found here.
Setup
In this example, we will be creating a repository for an analysis (R
script) stored in Integral that was built on version 3.3.6 of the
ggplot2 package. In the R code, there is an argument
size in the function geom_line() that became
deprecated in versions 3.4.0 and onward. Users of newer
ggplot2 versions can still run the script, but it returns
warnings in the console. In addition to striving for your analysis to
run with no warnings or errors, it is likely that this argument will
eventually be removed all together, meaning the analysis will not be
able to be run on later versions. Because setting up a working
environment for new users with outdated packages can be tedious to get
all the dependencies aligned, we will create a repository with the last
version of ggplot2 before the size argument
was deprecated, to help maintain reproducibility for all future
users.
To prepare this repository snapshot with the required R packages and versions we need, we will download our analysis script from Integral.
library(Certara.IntegralR)
integral_download(
file = "R Package Repository - Demo/R Scripts/pk_conc_time_plot.R",
path = getwd()
)Accessing Old Packages from CRAN
The next step is to install the the required version of
ggplot2 so we can build a lockfile for future users. The
Comprehensive R Archive Network (CRAN) is the main repository in which R
packages are stored and installed from. Packages can be installed in two
different ways:
- From “binary”. This is a ‘pre-compiled’ and
‘pre-built’
.zip(Windows) or.tgz(Mac) file of the package specific to your OS and R version. The contents of the binary file will be similar to the contents of the installed package folder inside your R library. Downloading binaries is the preferred option as the binary file simply needs to be extracted and moved inside your R library folder, e.g.,.libPaths(), making installation significantly faster.
- From “source”. These files are saved as
.tar.gzcompressed folders and contain the raw source code of the package. Installing from source is slower because, after the.tar.gzis downloaded, the package must then be built on the user’s system, which can take additional time. Further, if the particular R package calls C, C++, Fortran code internally or requires external libraries that need linking (e.g., BLAS or LAPACK), this will require additional compilation and RTools may be required. Linux does not commonly distribute R package binaries as precompiled files. Instead, users typically build packages from source (.tar.gz) and the required system libraries must be installed separately from the Linux terminal e.g.,sudo apt install libblas-dev liblapack-dev.
The most common way to install old package versions is by using the
remotes package. With the install_version()
function, you can install previous versions of packages from CRAN.
remotes::install_version("ggplot2", version = "3.3.6")While this will work, it is possible that the version you select will need compilation from source. CRAN only maintains binaries for the newest package versions that were available at the time of each minor R release (e.g., 4.3, 4.4). Additionally, they do not maintain binaries for any previous major R version (e.g., 3.0). In our case, our closest options for binaries are version 3.3.5 (R 4.0) or 3.4.2 (R 4.1), neither of which are the version we want.
Cases like this mean we would need to find a different repository than the central CRAN if we want this package binary. Establishing a repository of source files would be redundant for us since CRAN already hosts them. Additionally, it could force downstream users to compile the packages locally after installation, which can be a tedious process for re-running analyses and may lead to installation discrepancies or compilation issues.
Using CRAN Snapshots
Luckily, Posit (the creator of RStudio) has been taking various snapshots of the entire CRAN repository almost daily since 2017. These snapshots contain every package (source and binary) that was on CRAN at the time of snapshot, so while downloading the entire snapshot would be far too cumbersome for individual use, we can still download the specific packages we need.
Heading to https://packagemanager.posit.co/client/#/repos/cran/setup,
we can follow the prompts to gain access to the specific repository
required to download our packages. By selecting the date of November 3,
2022 (the most recent date before ggplot2 was updated to
version 3.4.0), we are given a URL similar to the traditional CRAN one,
but this is to the repository as it stood on that specific date. We will
pass this URL to all repo arguments to ensure that we are
pulling packages from the snapshot when ggplot2 was at the
desired version, increasing the likelihood of finding binaries.
We will download any packages needed for our analysis from this new
URL, to ensure compatibility and avoid introducing newer dependencies
that could disrupt the consistency of our environment. renv
will use the traditional CRAN URL, as newer versions have helper
functions and the package itself does not have any dependencies outside
of base-R packages. Because we are using IntegralR in our
workflow, we will also include the Certara repository URL when checking
for dependencies.
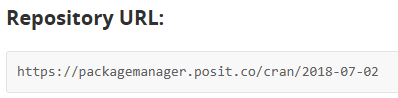
CRAN_url <- "https://packagemanager.posit.co/cran/2022-11-03"
# Install necessary packages
install.packages("renv")
install.packages("ggplot2", repos = CRAN_url)
install.packages("miniCRAN", repos = CRAN_url)
install.packages("Certara.IntegralR",
repos = c(Certara = "https://certara.jfrog.io/artifactory/certara-cran-release-public/",
CRAN = CRAN_url),
method = "libcurl")Create a Lockfile and Repo
With our CRAN repository URL, we can now start the process of
creating a temporary repository and its associated lockfile, as
demonstrated in the overview vignette. We will specify the repos we are
using in the snapshot() function so they can be recorded in
the lockfile.
# Create lockfile
renv::snapshot(repos = c(CRAN = CRAN_url,
Certara = "https://certara.jfrog.io/artifactory/certara-cran-release-public/"))
# Parse lockfile
lockfile <- renv::lockfile_read()
# Parse package names
pkgs <- names(lockfile$Packages)
# Get list of all recursive ggplot2 dependencies as of 2022-11-03
pkgList <- miniCRAN::pkgDep(pkgs,
repos = c(CRAN = CRAN_url,
Certara = "https://certara.jfrog.io/artifactory/certara-cran-release-public/"),
type = "source", suggests = FALSE)
# List of packages to be added to repository
pkgList [1] "Certara.IntegralR" "MASS" "Matrix" "R6" "RColorBrewer"
[6] "askpass" "cli" "colorspace" "curl" "digest"
[11] "fansi" "farver" "ggplot2" "glue" "gtable"
[16] "httr2" "isoband" "jsonlite" "labeling" "lattice"
[21] "lifecycle" "magrittr" "mgcv" "munsell" "nlme"
[26] "openssl" "pillar" "pkgconfig" "rappdirs" "renv"
[31] "rlang" "scales" "sys" "tibble" "utf8"
[36] "uuid" "vctrs" "viridisLite" "withr"
# Create temporary directory
repo_path <- file.path(tempdir(), "temp_repo")
dir.create(repo_path)
# Make a repository
miniCRAN::makeRepo(pkgList,
path = repo_path,
repos = c(CRAN = CRAN_url,
Certara = "https://certara.jfrog.io/artifactory/certara-cran-release-public/"),
type = c("source", "win.binary"))Modifying Our Repo
Sometimes after creating a repository, you may realize that you need to add more packages or update one that exists in the repository. miniCRAN provides a few helper functions to assist with this.
Updating a Package Version
If we decide that one of the packages needs updating, we can use
oldPackages() to return a list of the packages that have a
newer version available. Once we have decided that we want one of those
updated versions, we can use updatePackages(). This will
update those packages to the newer version, replacing the previous one
in our repo. The default behavior will interactively ask if you want to
update each package individually, but adding the
ask = FALSE argument can silence this and forcefully update
all of them. These functions can only update one type at a
time, so be sure to update all types equally to ensure your repository
does not have different versions within it. For this case, we need to
update the curl package to >= 5.0.1 to satisfy the
requirements for IntegralR. We will use the first date
available after version 5.0.1 was released.
# Check what packages have updates. Indexing [,1:3], as column 4 is just the repository URL
miniCRAN::oldPackages(path = repo_path, repos = "https://packagemanager.posit.co/cran/2023-06-08", type = "source")[,1:3]
miniCRAN::oldPackages(path = repo_path, repos = "https://packagemanager.posit.co/cran/2023-06-08", type = "win.binary")[,1:3] Package LocalVer ReposVer
cli "cli" "3.4.1" "3.6.1"
colorspace "colorspace" "2.0-3" "2.1-0"
curl "curl" "4.3.3" "5.0.1"
digest "digest" "0.6.30" "0.6.31"
fansi "fansi" "1.0.3" "1.0.4"
ggplot2 "ggplot2" "3.3.6" "3.4.2"
gtable "gtable" "0.3.1" "0.3.3"
httr2 "httr2" "0.2.2" "0.2.3"
isoband "isoband" "0.2.6" "0.2.7"
jsonlite "jsonlite" "1.8.3" "1.8.5"
lattice "lattice" "0.20-45" "0.21-8"
MASS "MASS" "7.3-58.1" "7.3-60"
Matrix "Matrix" "1.5-1" "1.5-4.1"
mgcv "mgcv" "1.8-41" "1.8-42"
nlme "nlme" "3.1-160" "3.1-162"
openssl "openssl" "2.0.4" "2.0.6"
pillar "pillar" "1.8.1" "1.9.0"
renv "renv" "0.16.0" "0.17.3"
rlang "rlang" "1.0.6" "1.1.1"
sys "sys" "3.4.1" "3.4.2"
tibble "tibble" "3.1.8" "3.2.1"
utf8 "utf8" "1.2.2" "1.2.3"
vctrs "vctrs" "0.5.0" "0.6.2"
viridisLite "viridisLite" "0.4.1" "0.4.2"
# Go ahead and update the curl package
miniCRAN::updatePackages(path = repo_path, oldPkgs = "curl",repos = "https://packagemanager.posit.co/cran/2023-06-08", type = "source", ask = FALSE)
miniCRAN::updatePackages(path = repo_path, oldPkgs = "curl",repos = "https://packagemanager.posit.co/cran/2023-06-08", type = "win.binary", ask = FALSE)While the repository is updated, our lockfile is not, as it is based
on the versions we have in our local library. We will need to update our
local version before we can update the lockfile. We will install
curl from the same CRAN snapshot date that was used in
making the repo.
# Update curl, using the newer CRAN snapshot date
install.packages("curl", repos = "https://packagemanager.posit.co/cran/2023-06-08", dependencies = FALSE)
# Update lockfile
renv::snapshot(repos = c(CRAN = CRAN_url,
Certara = "https://certara.jfrog.io/artifactory/certara-cran-release-public/"))Adding a New Package
We can also add entirely new packages to our repo using the
addPackage() function. Here we will add the
tictoc package in case downstream users want to run some
benchmark tests on the analysis. The default behavior downloads all
dependencies and suggests, so after checking there are
no dependencies we need, we will clarify the deps = FALSE
argument to prevent any unnecessary extra packages.
# Check for dependencies
miniCRAN::pkgDep("tictoc", repos = CRAN_url, type = "source", suggests = FALSE)
# Download and ignore dependencies/suggests
miniCRAN::addPackage("tictoc",
path = repo_path,
repos = CRAN_url,
deps = FALSE)Because we did not install this package locally and update our
lockfile, downstream users will not automatically have the
tictoc package installed when they restore their
environment. However, it will be available in their local repository,
allowing them to install the version we saved for them if they wish.
Using the Repository
At this stage, we would zip the repository and upload it to Integral as demonstrated in the overview vignette. The repository is now set up for future users to easily replicate the analysis. A downstream user would first want to move to a clean analysis folder and download the necessary lockfile, repository, and analysis script from Integral, making sure to unzip the repository into their working directory.
library(Certara.IntegralR)
# Download the repository and lockfile to the new folder
Certara.IntegralR::integral_download(
file = c("R Package Repository - Demo/R Scripts/pk_conc_time_plot.R",
"R Package Repository - Demo/Repository/renv.lock",
"R Package Repository - Demo/Repository/repo.zip"),
path = getwd()
)They then will want to activate and load their renv
before restoring it with the lockfile. Make sure to point to the new
unzipped repository in the repos argument.
# Activate and load renv
renv::activate()
renv::load()
# Restore renv from lockfile
renv::restore(repos = "repo")With everything set up, they should now be able to execute the script
and successfully recreate the figure using the deprecated arguments that
only work in ggplot2 version 3.3.6 and earlier.
library(Certara.IntegralR)
library(ggplot2)
pkData <- Certara.IntegralR::integral_read(FUN = read.csv,
file = "R Package Repository - Demo/Data/pkData.csv")
# Create plot of concentration vs. time
pkData$Subject <- as.factor(pkData$Subject)
pkData |>
ggplot(aes(x = Act_Time, y = Conc, group = Subject, color = Subject)) +
scale_y_log10() +
geom_line(size = 0.5) + # The size arg results in a warning in ggplot2 >= 3.4.0
geom_point() +
ylab("Drug Concentration \n at the central compartment")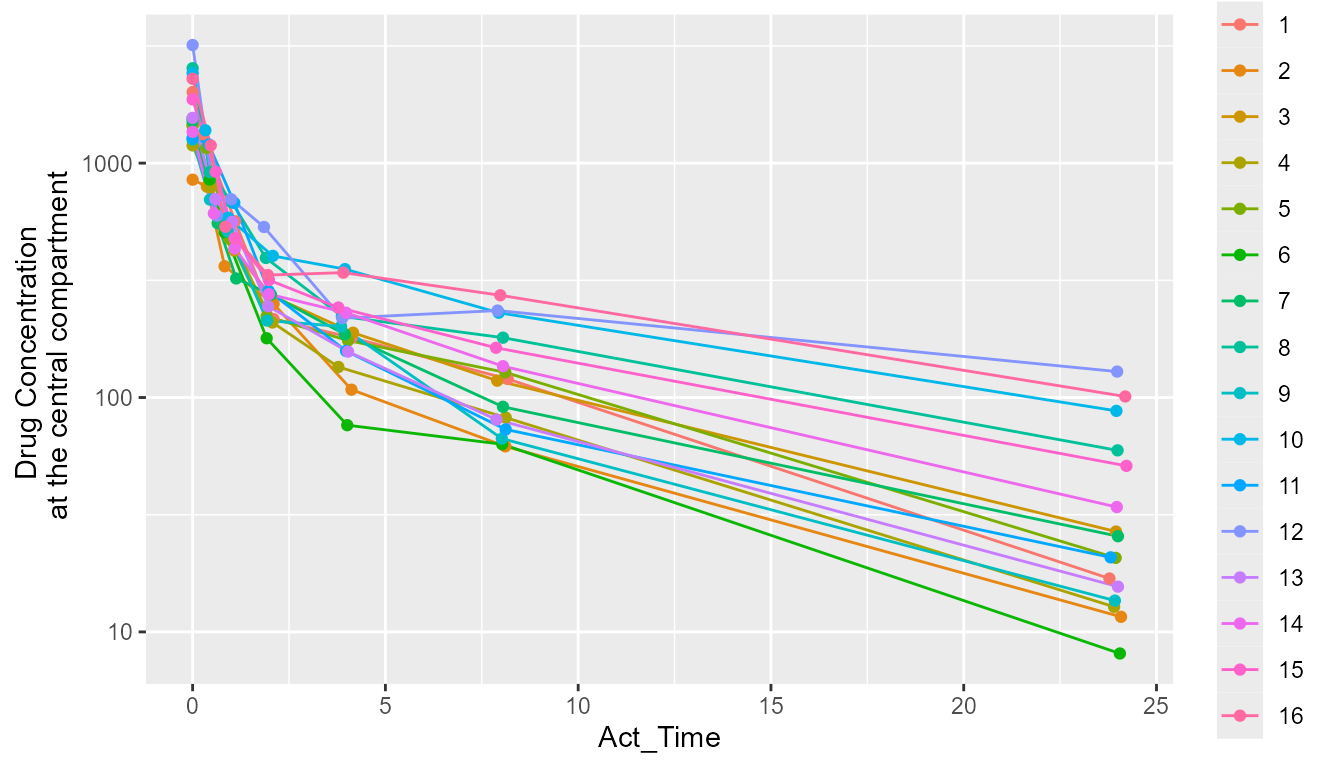
Troubleshooting
Mismatched Repo and Lockfile
If restoring from your lockfile results in errors that the specified
versions cannot be found, it is likely that your local package library,
when you created the lockfile, did not have the packages versions you
intended. renv::snapshot() checks for required packages
that are called in R scripts, but the versions it stores are the ones
that were present in your local library. It is common for local versions
to be newer than those that were obtained using a CRAN snapshot URL. The
easiest way to solve this is to erase the entire local library and start
from scratch, installing only the required packages to have the scripts
run, and sourcing them all from the SAME repository
that you are downloading from in your makeRepo() call.
Additionally, make sure you are not accidentally downloading any
packages outside of the required dependencies. Recursive dependencies
can become complex, so you only want to download the bare minimum to
reproduce the analysis.
Duplicate Packages in Repo
The default behavior of addPackage() will add
duplicate versions of dependency packages to your repo,
leaving it in an inconsistent state. The easiest way to avoid this is to
add the deps = FALSE argument to the
addPackage() function, and download each needed package on
its own. This can become very complicated when the list of recursive
dependencies is large, and is the reason why setting up the repo
correctly in a single call to makeRepo(), ideally with the
newest version of packages, is the cleanest and simplest way to build
your repository. If you realize you need a new package and are
encountering issues with adding it to the miniCRAN repo, it
may be worth restarting and making sure you include it in the first call
to makeRepo().
If you encounter this issue and choose to move forward, you will need to manually check the repository for duplicates and remove them before updating the PACKAGES index.
Below is an example of how this may happen, and how to resolve it.
# List packages
pkgs <- miniCRAN::pkgDep("askpass", repos = CRAN_url, type = "source", suggests = FALSE)
# Make a repository
my_dir <- tempdir()
miniCRAN::makeRepo(pkgs,
path = my_dir,
repos = c(CRAN = CRAN_url),
type = c("source", "win.binary"))
# Add a new packages from the current CRAN
# Not specifying deps = FALSE will add some newer versions of dependencies
miniCRAN::addPackage("openssl",
path = my_dir,
repos = "https://cloud.r-project.org")
# List packages, and return warning if there are duplicates
src_pkgs <- miniCRAN::checkVersions(pkgList, path = my_dir, type = "source")
bin_pkgs <- miniCRAN::checkVersions(pkgList, path = my_dir, type = "win.binary")We get the following warning returned when checking our source packages, but not the binaries.
Warning message:
Duplicate package(s): askpass, sys We then need to manually index the source packages and remove the duplicates.
# After inspecting package versions, remove old versions
basename(src_pkgs[[1]]) [1] "R6_2.5.1.tar.gz" "askpass_1.1.tar.gz" "askpass_1.2.1.tar.gz" "cli_3.6.3.tar.gz" "curl_6.0.1.tar.gz"
[6] "digest_0.6.37.tar.gz" "glue_1.8.0.tar.gz" "jsonlite_1.8.9.tar.gz" "lifecycle_1.0.4.tar.gz" "magrittr_2.0.3.tar.gz"
[11] "openssl_2.3.0.tar.gz" "rappdirs_0.3.3.tar.gz" "rlang_1.1.4.tar.gz" "sys_3.4.1.tar.gz" "sys_3.4.3.tar.gz"
[16] "withr_3.0.2.tar.gz"
# Remove the packages by index. Alternatively, you can supply the full file name
file.remove(src_pkgs[[1]][c(2, 14)])
# Update PACKAGES index
miniCRAN::updateRepoIndex(repo_path, type = c("source", "win.binary"))