Paliperidone Palmitate Case Study
paliperidone_example.RmdOverview
This Paliperidone Palmitate case study demonstrates a comprehensive Virtual Bioequivalence (VBE) analysis using the integrated Pirana-SimcypTM workflow. This study stems from an FDA grant and showcases the complete VBE process from initial setup through final statistical analysis.
The case study evaluates bioequivalence between reference and test formulations of Paliperidone Palmitate, focusing on the Critical Quality Attribute (CQA) of particle size distribution (PSD) mean radius. The analysis demonstrates how varying PSD parameters affects bioavailability and bioequivalence outcomes.
Study Design
Critical Quality Attribute (CQA)
The primary CQA for this study is the Particle Size Distribution (PSD) Mean Radius. This parameter directly influences drug dissolution and absorption, making it critical for bioequivalence assessment.
Formulation Parameters
| Parameter | Reference Value | Test Range | Units |
|---|---|---|---|
| PSD Mean Radius | 4.6357 | 1.80 - 7.50 | µm |
Virtual Population Design
The study includes one reference formulation and multiple test formulations with varying PSD mean radius values:
| PSD Mean Radius | Type | Number of Subjects | Description |
|---|---|---|---|
| 4.6357 | Reference | 100,000 | Reference formulation |
| 1.8000 | Test | 2,500 | Lower bound test |
| 2.0000 | Test | 2,500 | Lower bound test |
| 2.6000 | Test | 2,500 | Lower bound test |
| 2.7762 | Test | 50,000 | Type I error boundary |
| 3.0000 | Test | 50,000 | Lower range test |
| 4.6357 | Test | 40,000 | Perfect bioequivalence test |
| 4.8375 | Test | 40,000 | Upper range test |
| 5.0679 | Test | 40,000 | Upper range test |
| 5.5000 | Test | 100,000 | Upper range test |
| 6.2300 | Test | 100,000 | Upper range test |
| 6.4500 | Test | 100,000 | Upper range test |
| 6.7000 | Test | 2,500 | Upper bound test |
| 6.8230 | Test | 50,000 | Type I error boundary |
| 7.1000 | Test | 2,500 | Upper bound test |
| 7.5000 | Test | 20,000 | Upper bound test |
Trial Design
The case study uses a parallel trial design. Currently, only parallel VBE trial designs are supported in the Pirana-Simcyp VBE module.
Simulation Performance
Estimated simulation run times across different subject numbers and available cores:
| Number of Subjects | Number of Cores | Estimated Time (minutes) | Estimated Time (hours) |
|---|---|---|---|
| 1,000 | 8 | 53.08 | 0.88 |
| 1,000 | 16 | 26.54 | 0.44 |
| 1,000 | 32 | 13.27 | 0.22 |
| 10,000 | 8 | 530.79 | 8.85 |
| 10,000 | 16 | 265.39 | 4.42 |
| 10,000 | 32 | 132.70 | 2.21 |
| 100,000 | 8 | 5,307.87 | 88.46 |
| 100,000 | 16 | 2,653.94 | 44.23 |
| 100,000 | 32 | 1,326.97 | 22.12 |
Setup and Configuration
Prerequisites
Note: Make sure you have completed Setup and Configuration before proceeding with this tutorial.
Example Files
Click here to download the example Pirana VBE workspace files for Paliperidone Palmitate.
The archive contains:
- Example
vbefiles and simulation results files (csv) - 1 reference
vbeobject and 15 testvbeobjects - Complete simulation outputs across different CQA values
Please download and unzip the file archive and navigate to the resulting PP3 folder in your Pirana working directory.
Important Note: Pre-Generated Simulation Results
All simulation results have been pre-generated and provided with the example files. This means:
- Do NOT re-run existing VBEs - The example VBEs already contain complete simulation results
- Use the provided results - You can run R scripts directly on the existing simulation data
- Avoid data overwriting - Re-running simulations would overwrite the original results in your working directory
- Create new VBEs for additional simulations - If you want to generate new simulation results, create new VBE objects with different names
The example files are designed to allow you to explore the R script functionality immediately without waiting for simulations to complete. You can run through all the statistical analysis scripts and generate outputs to understand the workflow.
Working Directory Setup
- Navigate to your Pirana working directory where the example files have been copied
-
Select the VBE context in Pirana and ensure
All foldersis selected - Explore VBE objects clicking a vbe model object in the Pirana table, you can see simulation output subfolders highlighted from previous runs, with the latest simulation results copied from the subfolder into the current working directory.
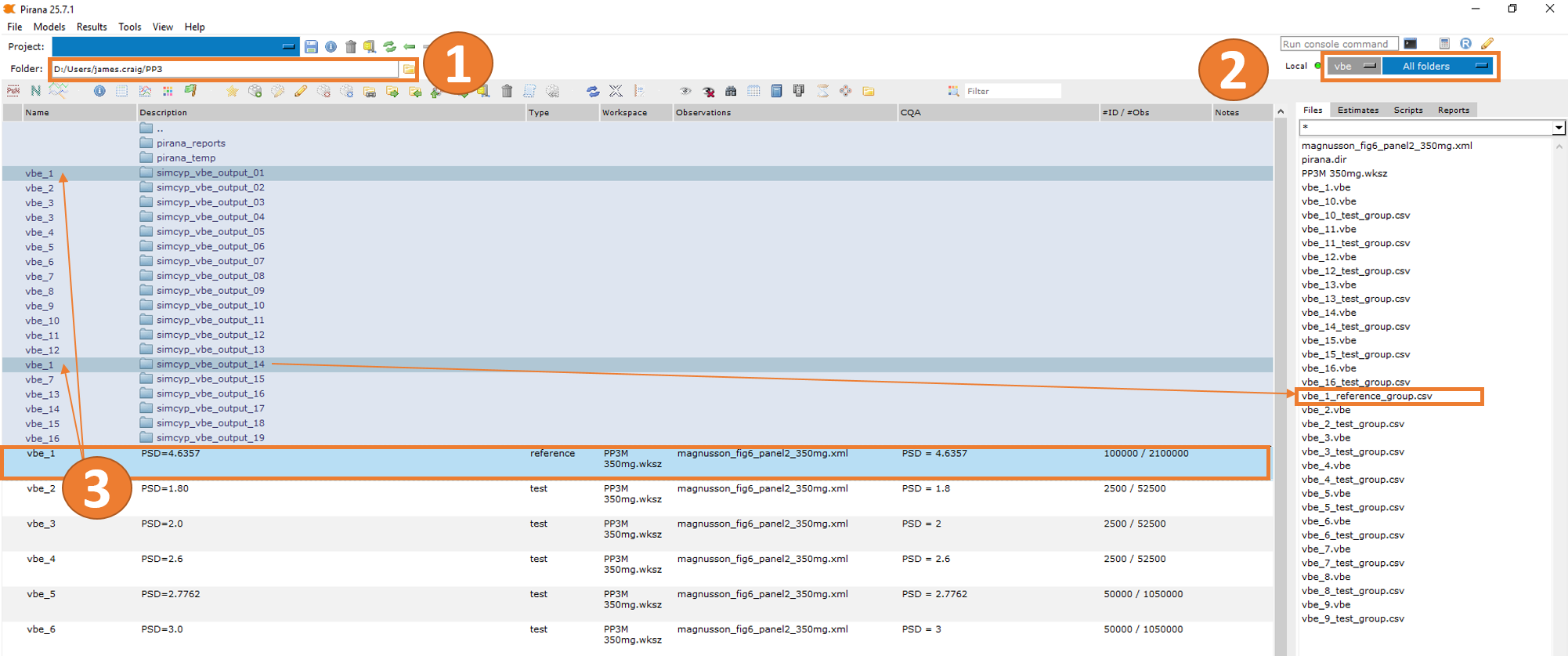
File Organization
When you select vbe_1 in the Pirana table, you’ll see
that SimcypTM simulation output subfolders (e.g.,
simcyp_vbe_output_01 and simcyp_vbe_output_14)
are “attached” to the VBE object (as illustrated in item 3 in the figure
above).
These subfolders contain the simulation results from the
SimcypTM simulations executed in the Shiny application. Each
time a new Shiny session is launched and a simulation is run for the
selected vbe, upon exiting the Shiny session, the most
recent simulation file generated is automatically copied from the
simulation output folder to the Pirana working directory and renamed to
{vbe_name}_{vbe_type}_group.csv (e.g.,
vbe_1_reference_group.csv).
Note: To keep the example file size manageable, the simulation
output subfolders contain only a subset of subjects from the full
simulation runs. Complete simulation results with all subjects are
available in the corresponding .csv files located in the Pirana working
directory (e.g., vbe_1_reference_group.csv).
Phase 1: Reference Group Simulation (Optional)
Note: Since simulation results are pre-generated in the example files, you can skip Phases 1-3 and proceed directly to Phase 4: Statistical Analysis if you want to explore the R script functionality immediately.
Step 1: Create Reference VBE
- Navigate to the top-level menu in Pirana
- Select Models > New Model (or use the shortcut
Ctrl + n) - This will launch the ‘Create VBE’ dialog
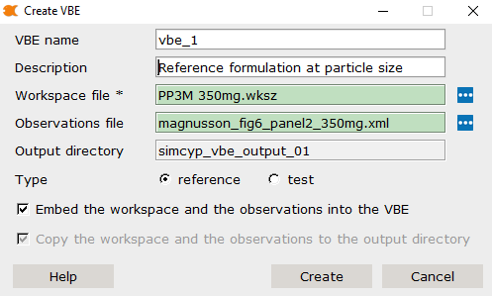
Step 2: Configure Reference VBE Settings
VBE Configuration
-
VBE Name:
vbe_1(default) - Description: “Paliperidone Palmitate Reference Formulation”
- Type: Reference
File Selection
-
Workspace File:
PP3M 350mg multidose June 24 variability calibrated model newseed.wksz -
Observations File:
magnusson_fig6_panel2_350mg.xml
This workspace file contains calibrated input parameters for the
reference group formulation with the Particle Size Distribution Mean
Radius parameter value set to 4.6357.
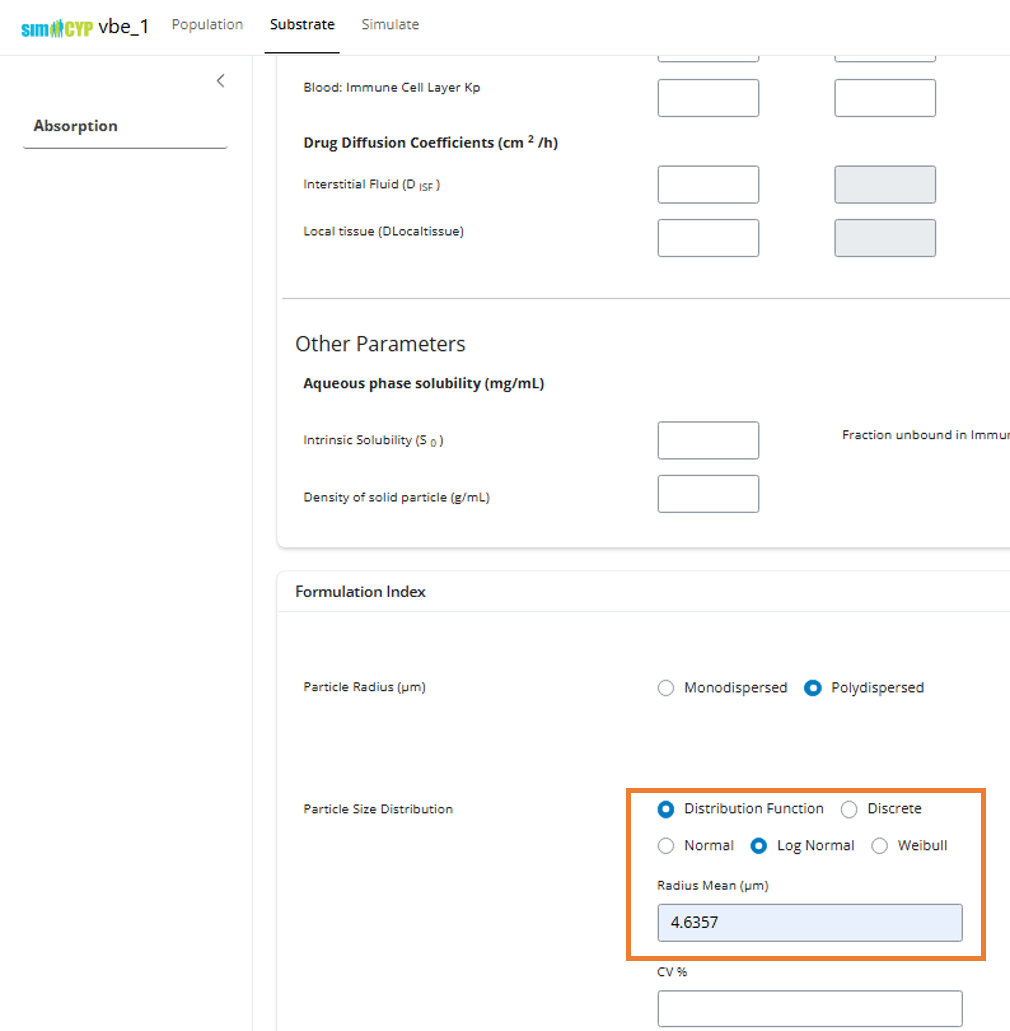
Step 3: Set Reference Parameters
- Navigate to the Formulation Index section under Substrate > Absorption
- Set the PSD Mean Radius parameter value to
4.6357
Step 4: Run Reference Simulation
- Navigate to the Simulate tab in the top menu
- Click the Simulate button in the left sidebar
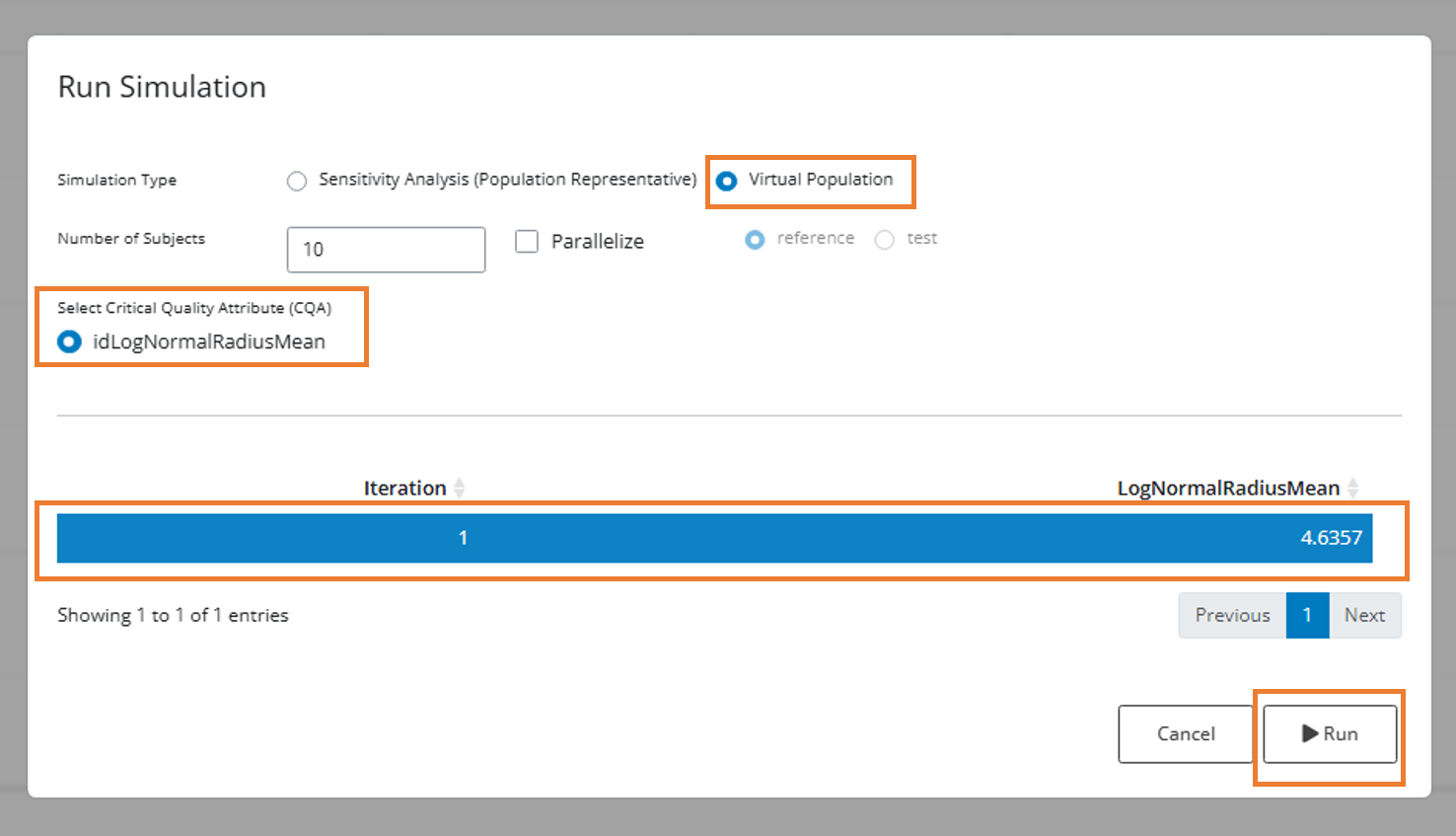
- Configure the simulation settings:
- Virtual Population: Select this radio button
- Number of Subjects: 100,000
- Parallelize: Check this box for faster simulation
- Parameter Table: Select the row with PSD Mean Radius = 4.6357
Phase 2: Sensitivity Analysis (Optional)
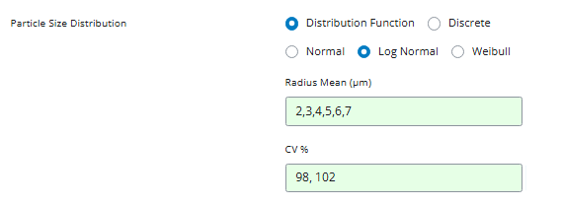
Step 1: Set Multiple Parameter Values
Return to the parameter input that you set for the reference group. For sensitivity analysis, you’ll want to test multiple PSD values around the reference value.
The application supports three input formats:
-
Single value:
4.6357 -
Comma-separated list:
1.8000, 2.0000, 2.6000, 2.7762, 3.0000, 4.6357, 4.8375, 5.0679, 5.5000, 6.2300, 6.4500, 6.7000, 7.1000, 7.5000 -
Sequence statement:
seq(1.8, 7.5, 0.5)(sequence from 1.8 to 7.5 by 0.5)
For this example, let’s test values around our reference value of
4.6357:
Step 2: Preview Parameter Combinations
Navigate to the Simulate tab and select Parameters. You’ll see an expanded grid showing all unique parameter combinations given the input values we’ve set:
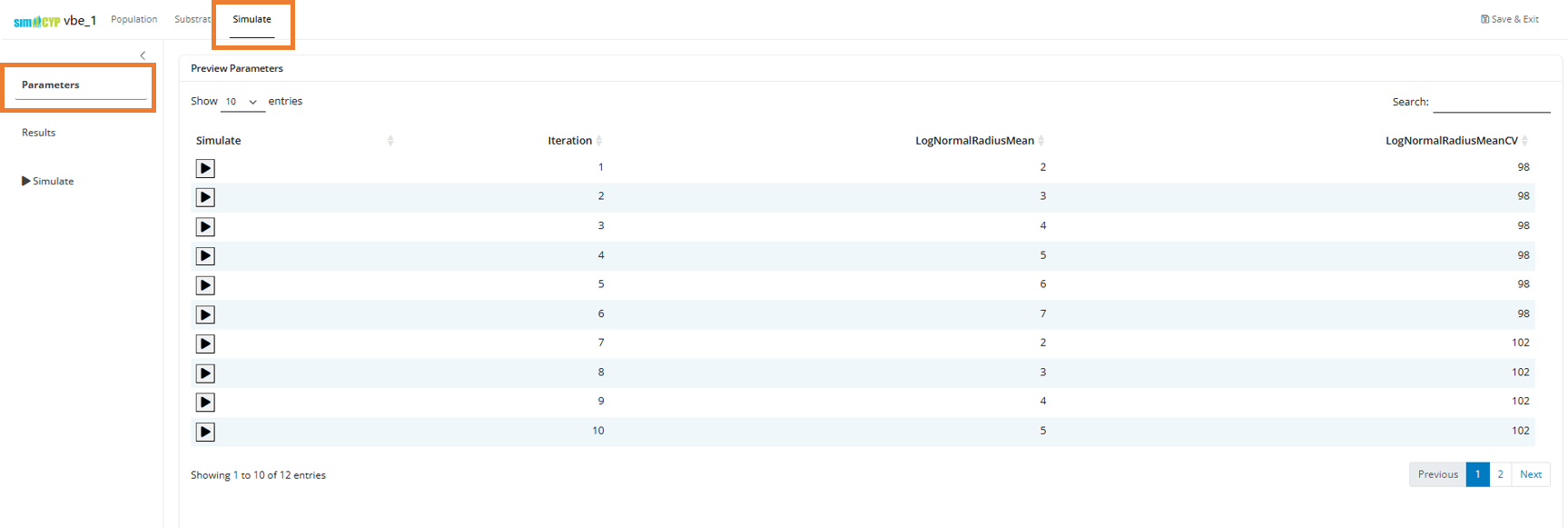
The iteration number corresponds to the unique identifier for each parameter combination.
Step 3: Run Sensitivity Analysis
Instead of simulating individual parameter combinations, select the “Sensitivity Analysis (Population Representative)” radio button. This will perform simulations for a representative subject in the population for each unique parameter combination.
Step 4: Analyze Sensitivity Results
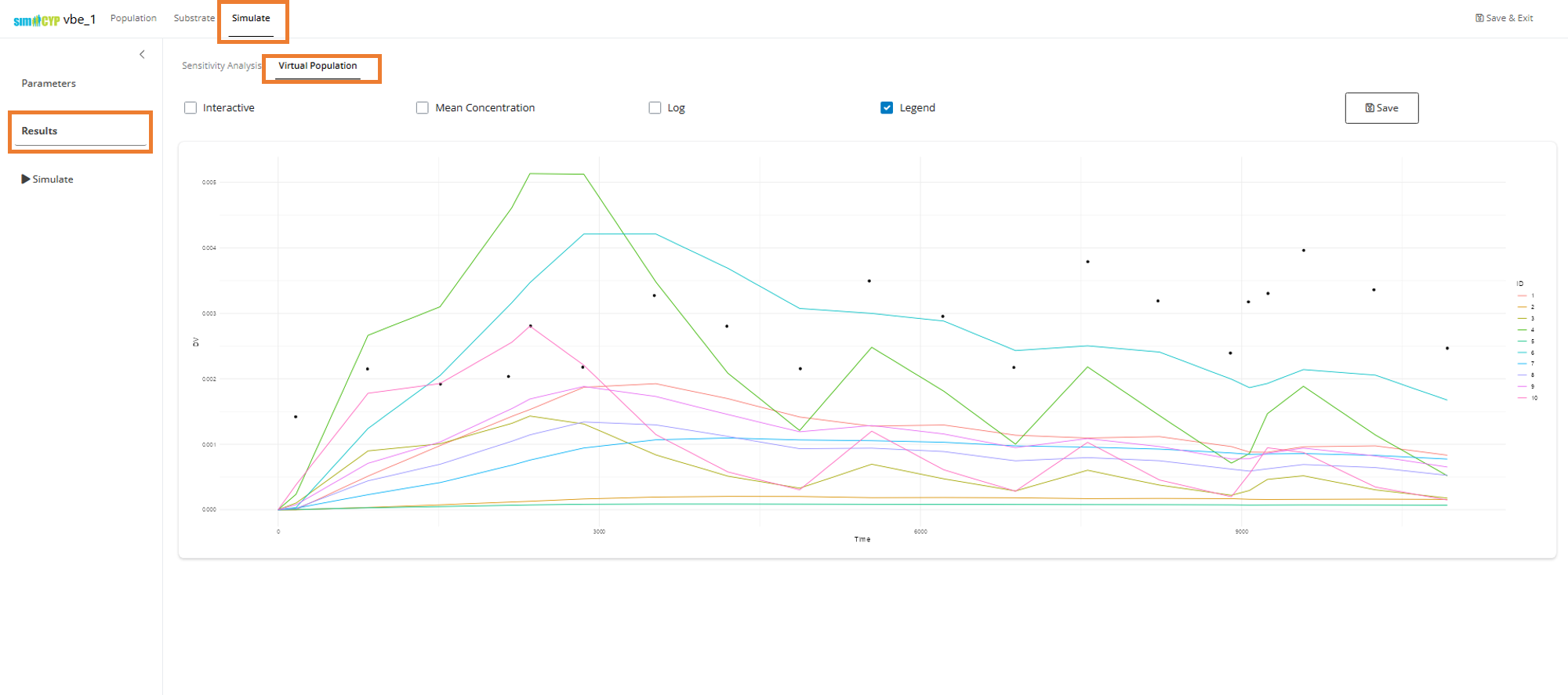
Navigate to the Results tab and select Sensitivity Analysis to view your sensitivity analysis results:
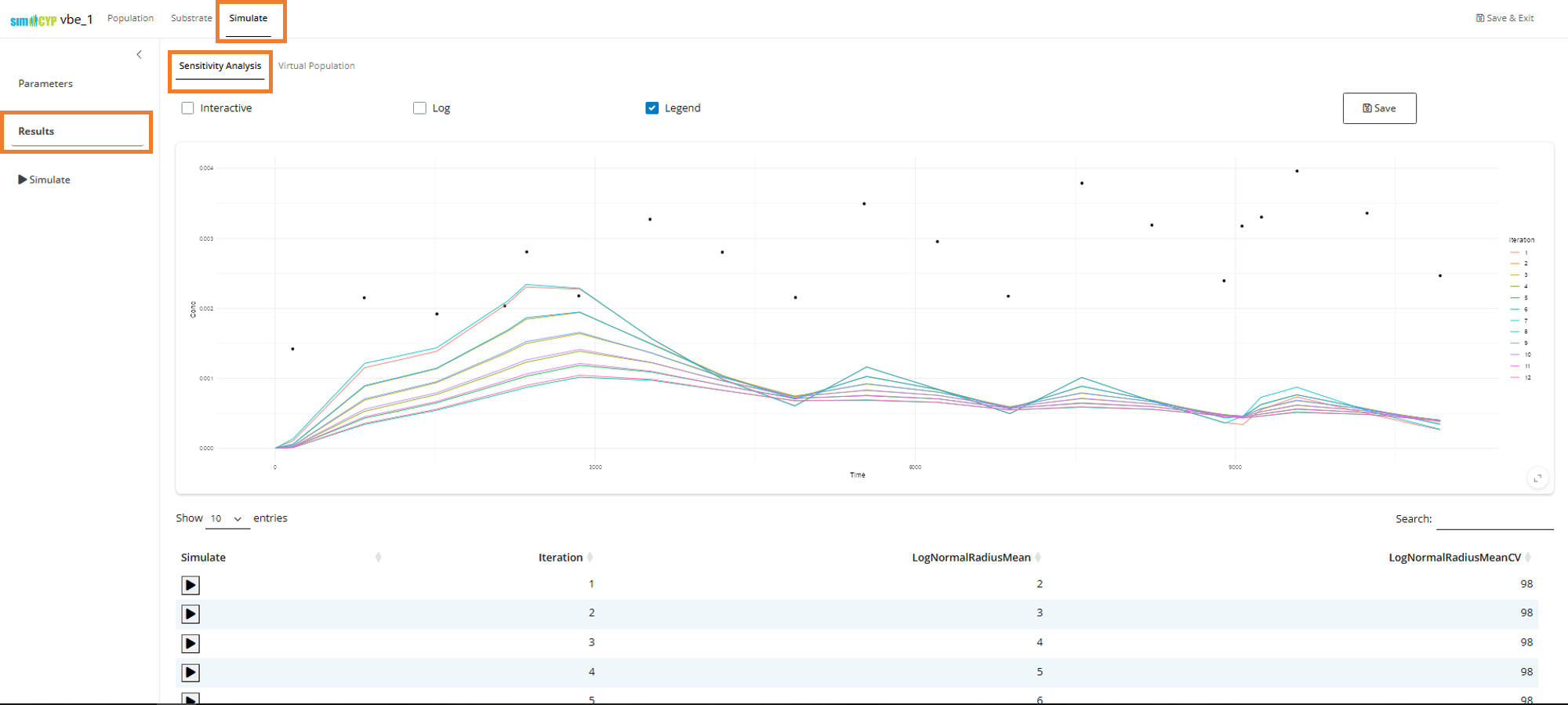
You can select any row in the table to observe the best fit through the data.
Note: Because the drug has very high variance and the distributions are log-normal, we’re seeing that the observed median value (points) is higher than the population representative predictions (lines) returned in the above sensitivity analysis plot.
Phase 3: Test Group Simulations (Optional)
Based on your sensitivity analysis, you can now identify the CQA values you want to test for bioequivalence. You will need to create a separate VBE for each test CQA value you want to evaluate.
This process is iterative. You don’t need to create all VBEs upfront. Instead, you typically:
- Start with a single test CQA value and simulate a certain number of subjects
- Run R scripts to compare test results against your reference
- Check bioequivalence statistics
- Adjust simulations as needed (increase subjects, modify parameters)
- Repeat the process until you achieve the desired statistical power
Creating Test VBEs
For each test CQA value you want to evaluate:
- Create a new VBE using the same steps as for the reference group
(using the same
.wksz) - In the Create VBE dialog, select the ‘test’ option instead of ‘reference’
- Set the specific CQA value for this test formulation
- Run the virtual population simulation with the appropriate number of subjects
- Compare results with your reference group
Phase 4: Statistical Analysis
Once you have completed simulations for both reference and test groups, you can perform comprehensive statistical analysis using the integrated R scripts in Pirana.
Available Analysis Scripts
The VBE R script library is located in the Pirana installation folder
(e.g., {Pirana_Installation_Directory}/scripts/VBE). With
the VBE context selection selected in Pirana, the scripts will
automatically be visible in the scripts tab.
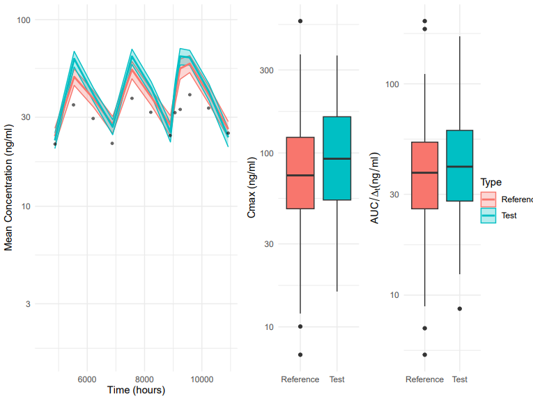
1. Compare Simulations
These scripts allow users to plot the time-concentration profiles with confidence intervals and additional plots for comparing AUC and CMax. Users can supply time filtering values to calculate AUC and CMax in a specific dosing region.
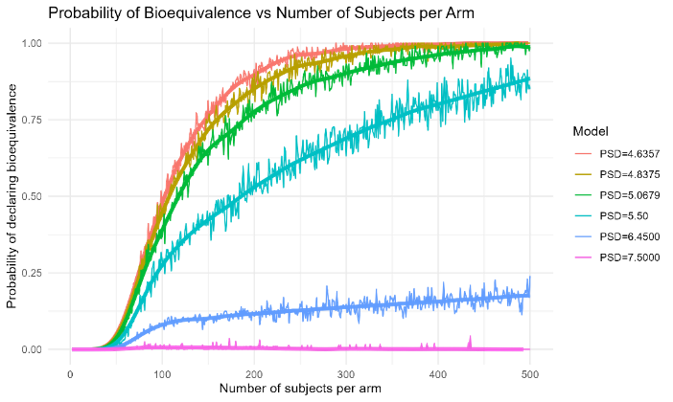
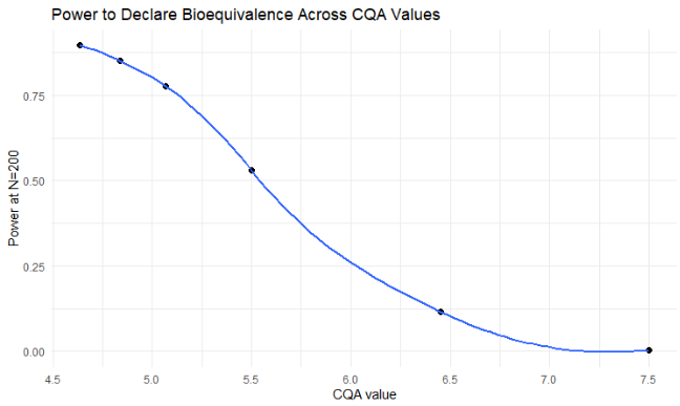
2. Power Curve vs Sample Size Analysis
This functionality estimates the statistical power (i.e., the probability of declaring bioequivalence) of TOST test assessments for VBE trials of varying sizes. A bootstrap Monte Carlo resampling algorithm is used to form VBE trials of different arm sizes from a large pool of simulated patients.
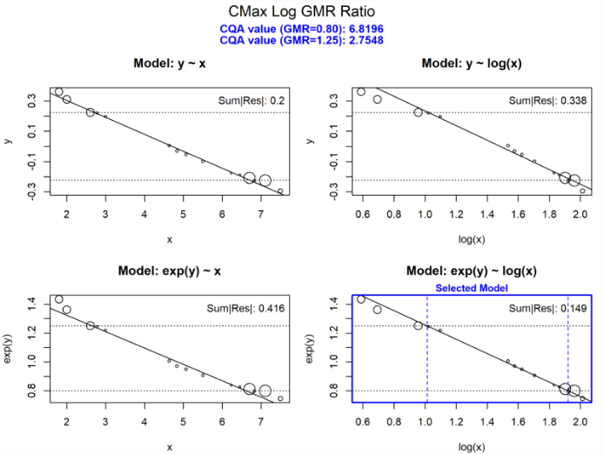
3. CQA Sensitivity Analysis
The cqa_sensitivity.R script evaluates the relationship
between a CQA and the geometric mean ratio (GMR) of either Cmax or AUC
in VBE simulations. It computes subject-level Cmax and AUC values from
reference and test datasets, optionally adding log-normal noise derived
from a user-defined sigma or estimated from observed data.
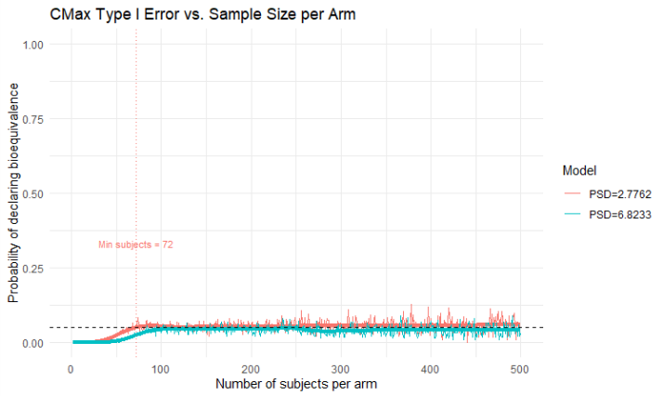
4. Type 1 Error Analysis
Estimates the type 1 error by calculating the probability of declaring bioequivalence for test drugs that are on the edge of the 0.8 - 1.25 GMR range.
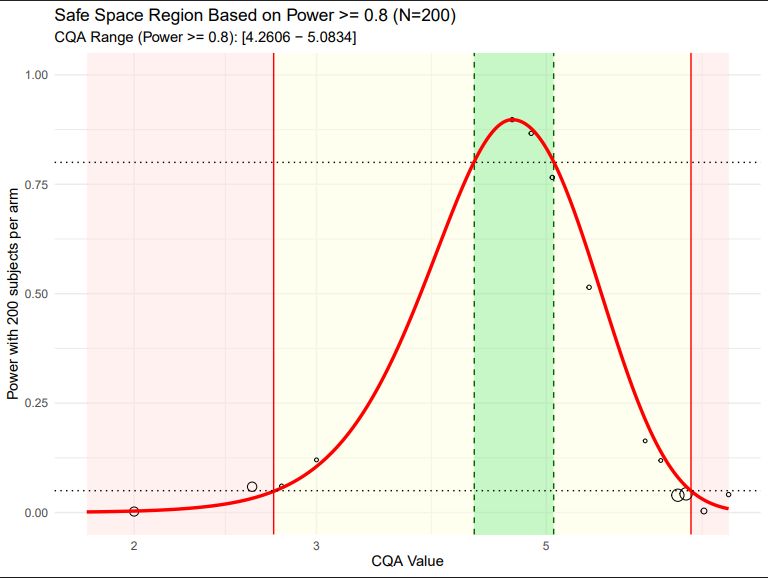
5. Safe Space Analysis
This script generates a plot of the safe space and will return the CQA bounds of this region to the user. Within this visualization, the region in which the population level GMRs are within the 0.8 to 1.25 range (as informed by the CQA sensitivity analysis) is highlighted to users.
Running Statistical Analysis
- Select your VBE objects in Pirana (reference and test VBEs, use ctrl/shift for multi-selection)
- Navigate to the Scripts tab in Pirana
- Choose the appropriate analysis script based on your objectives
- Configure script parameters as needed
- Execute the analysis and review results
Results and Interpretation
Key Findings
- Power Analysis: A trial size of 200 subjects per arm was determined to provide 85% power for the 4.8 µm radius test case
- Type I Error: The analysis correctly maintained type I error around the 5% level across increasing trial sizes
- Safe Space: The analysis identified the safe space boundaries where test formulations maintain bioequivalence with adequate statistical power
Statistical Power
The power analysis revealed that: - Smaller trial sizes (e.g., 50 subjects per arm) provide insufficient power for most test formulations - Larger trial sizes (e.g., 200+ subjects per arm) provide adequate power for formulations within the bioequivalence region - The relationship between CQA values and power is non-linear, with power decreasing rapidly as formulations move away from the reference
Type I Error Control
The type I error analysis demonstrated that: - Formulations at the bioequivalence boundaries (GMR = 0.8 and 1.25) correctly maintain type I error around 5% - The statistical methodology properly controls for false positive declarations of bioequivalence - The analysis provides confidence in the robustness of the VBE approach
Conclusions
This Paliperidone Palmitate case study demonstrates the comprehensive capabilities of the Pirana-SimcypTM VBE workflow. The analysis successfully:
- Established a robust reference simulation with 100,000 subjects
- Performed systematic sensitivity analysis across multiple CQA values
- Generated test simulations for multiple formulations
- Conducted comprehensive statistical analysis including power, type I error, and safe space assessment
- Provided actionable results for formulation development and quality control
The iterative nature of the VBE process allows researchers to refine their analysis based on preliminary results, making it a powerful tool for bioequivalence assessment in drug development.