Using Model Builder in RStudio for Textual Models
phoenix_mdl.RmdIn the following examples, you will learn how to:
- Read the input dataset and mdl file (a file for PML model with
.mdlextension) into RStudio - Use the
textualModel()function fromCertara.RsNLMEto create a textual model object in your R environment - Launch the textual model editor using
modelTextualUI(), map required columns, and specify covariate levels/labels
Data and model
The mdl and data, covariate.mdl and
covariate.csv, are from Certara University’s (201-OD)
Intermediate Population Modeling using Phoenix NLME. Both example files
are distributed with the Certara.RsNLME.ModelBuilder
package.
Import data
dataPath <- system.file("extdata", "covariate_data.csv", package = "Certara.RsNLME.ModelBuilder")
data <- read.csv(dataPath)
str(data)
#> 'data.frame': 511 obs. of 11 variables:
#> $ ID : int 1001 1001 1001 1001 1001 1001 1001 1001 1001 1001 ...
#> $ PT : int 1001 1001 1001 1001 1001 1001 1001 1001 1001 1001 ...
#> $ TIME : num 0 0.0486 0.0729 0.142 6.92 13.9 20.9 23 55 56.1 ...
#> $ CONC : num 0 6.11 5.59 5.12 2.37 ...
#> $ DOSE : num 25410 0 0 0 0 ...
#> $ RATE : int 0 0 0 0 0 0 0 0 0 0 ...
#> $ E : int 1 0 0 0 0 0 0 0 0 1 ...
#> $ M : int 1 0 0 0 0 0 0 0 0 1 ...
#> $ CMT : int 2 2 2 2 2 2 2 2 2 2 ...
#> $ WEIGHT: num 77 77 77 77 77 77 77 77 77 77 ...
#> $ SEX : chr "male" "male" "male" "male" ...Import .mdl
mdlPath <- system.file("extdata", "covariate.mdl", package = "Certara.RsNLME.ModelBuilder", mustWork = TRUE)
mdl <- readLines(mdlPath)
cat(gsub(pattern = "\t", "\n\t", mdl))
#> test(){
#> deriv(A1 = - Cl * C - Cl2 * (C - C2))
#> deriv(A2 = Cl2 * (C - C2))
#> dosepoint(A1, idosevar = A1Dose)
#> C = A1 / V
#> C2 = A2 / V2
#> error(CEps = 0.1)
#> observe(CObs = C * (1 + CEps))
#> stparm(V = tvV * exp(nV))
#> stparm(V2 = tvV2 * exp(nV2))
#> stparm(Cl = tvCl * exp(nCl))
#> stparm(Cl2 = tvCl2 * exp(nCl2))
#> fcovariate(WEIGHT)
#> fcovariate(SEX())
#> fixef(tvV = c(, 3739, ))
#> fixef(tvV2 = c(, 2852, ))
#> fixef(tvCl = c(, 210, ))
#> fixef(tvCl2 = c(, 897, ))
#> secondary(Ke = tvCl/tvV)
#> secondary(K12 = tvCl2/tvV)
#> secondary(K21 = tvCl2/tvV2)
#> secondary(r1 = ((K12+K21+Ke)^2-4*K21*Ke)^0.5)
#> secondary(Alpha = 0.5*(K12+K21+Ke+r1))
#> secondary(Beta = 0.5*((K12+K21+Ke)-r1))
#> secondary(A = (A1Dose/tvV*(Alpha-K21))/(Alpha-Beta))
#> secondary(B = (-(A1Dose/tvV)*(Beta-K21))/(Alpha-Beta))
#> secondary(AUC = A/Alpha+B/Beta)
#> secondary(Cmax = A1Dose/tvV)
#> secondary(AUMC = A/(Alpha*Alpha)+B/(Beta*Beta))
#> secondary(MRT = AUMC/AUC)
#> secondary(VSS = tvCl*MRT)
#> secondary(Ke_hl = log(2)/Ke)
#> secondary(Alpha_hl = log(2)/Alpha)
#> secondary(Beta_hl = log(2)/Beta)
#> ranef(diag(nV, nCl, nV2, nCl2) = c(1, 1, 1, 1)) }Create model object
Using the data in our R environment and the path to our
.mdl file, we can create a textual model object inside
RStudio using the textualmodel()
function from the Certara.RsNLME package, then pass that
model to the model argument of the
modelTextualUI() function from
Certara.RsNLME.ModelBuilder.
model <- textualmodel(modelName = "covariate", data = data, mdl = mdlPath)
#> Warning in checkCatCovariateMappingColumn(map, inputData, foundColumn,
#> modelTermName@variableName): The corresponding data column SEX for covariate
#> SEX is of class character. Please specify the name and the associated value for
#> each category through 'addLabel' function
print(model)
#>
#> Model Overview
#> -------------------------------------------
#> Model Name : covariate
#> Working Directory : C:/Users/mtalley/GitHub/R-RsNLME-model-builder/vignettes/covariate
#> Model Type : Textual
#>
#> PML
#> -------------------------------------------
#> test(){
#> deriv(A1 = - Cl * C - Cl2 * (C - C2))
#> deriv(A2 = Cl2 * (C - C2))
#> dosepoint(A1, idosevar = A1Dose)
#> C = A1 / V
#> C2 = A2 / V2
#> error(CEps = 0.1)
#> observe(CObs = C * (1 + CEps))
#> stparm(V = tvV * exp(nV))
#> stparm(V2 = tvV2 * exp(nV2))
#> stparm(Cl = tvCl * exp(nCl))
#> stparm(Cl2 = tvCl2 * exp(nCl2))
#> fcovariate(WEIGHT)
#> fcovariate(SEX())
#> fixef(tvV = c(, 3739, ))
#> fixef(tvV2 = c(, 2852, ))
#> fixef(tvCl = c(, 210, ))
#> fixef(tvCl2 = c(, 897, ))
#> secondary(Ke = tvCl/tvV)
#> secondary(K12 = tvCl2/tvV)
#> secondary(K21 = tvCl2/tvV2)
#> secondary(r1 = ((K12+K21+Ke)^2-4*K21*Ke)^0.5)
#> secondary(Alpha = 0.5*(K12+K21+Ke+r1))
#> secondary(Beta = 0.5*((K12+K21+Ke)-r1))
#> secondary(A = (A1Dose/tvV*(Alpha-K21))/(Alpha-Beta))
#> secondary(B = (-(A1Dose/tvV)*(Beta-K21))/(Alpha-Beta))
#> secondary(AUC = A/Alpha+B/Beta)
#> secondary(Cmax = A1Dose/tvV)
#> secondary(AUMC = A/(Alpha*Alpha)+B/(Beta*Beta))
#> secondary(MRT = AUMC/AUC)
#> secondary(VSS = tvCl*MRT)
#> secondary(Ke_hl = log(2)/Ke)
#> secondary(Alpha_hl = log(2)/Alpha)
#> secondary(Beta_hl = log(2)/Beta)
#> ranef(diag(nV, nCl, nV2, nCl2) = c(1, 1, 1, 1))
#> }
#>
#> Structural Parameters
#> -------------------------------------------
#> V V2 Cl Cl2
#> -------------------------------------------
#> Column Mappings
#> -------------------------------------------
#> Model Variable Name : Data Column name
#> id : ID
#> time : TIME
#> A1 : ?
#> WEIGHT : WEIGHT
#> SEX : SEX
#> CObs : ?We can see that the columns in our data that share the same name as
model variables get automatically mapped. We also received a warning
about required covariate labels for our categorical covariate
SEX, which is of class chr in the data.
unique(data$SEX)
#> [1] "male" "female"Map variables to columns through textual model builder
We can use the modelTextualUI() function to map the
remaining columns and add corresponding levels/labels for our
categorical covariate SEX.
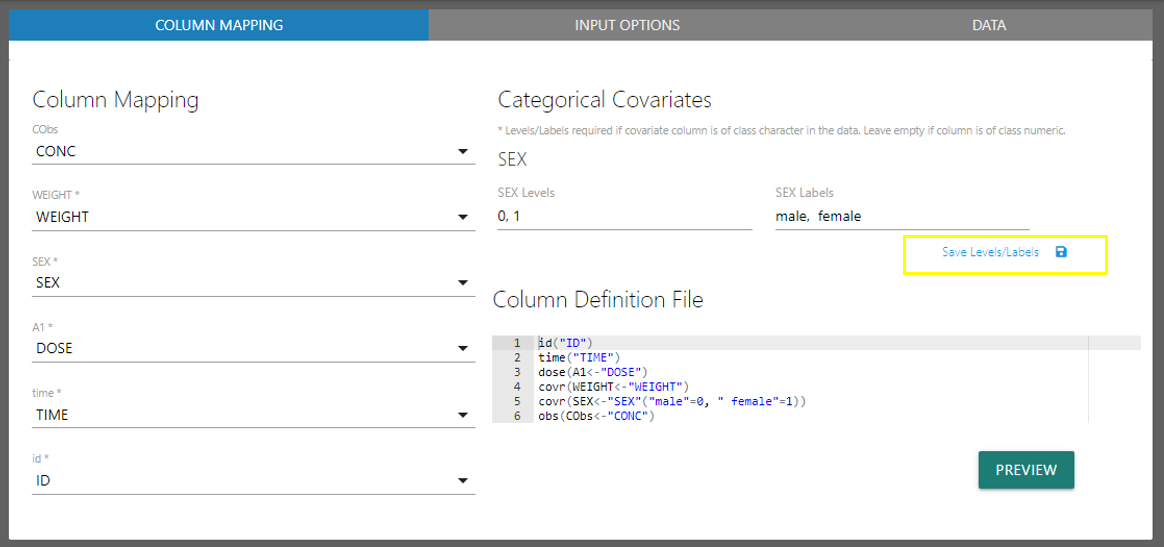
model <- modelTextualUI(model)After initializing the Shiny interface, we can see the PML editor on the left of the page, and various options related to column mapping on the right.
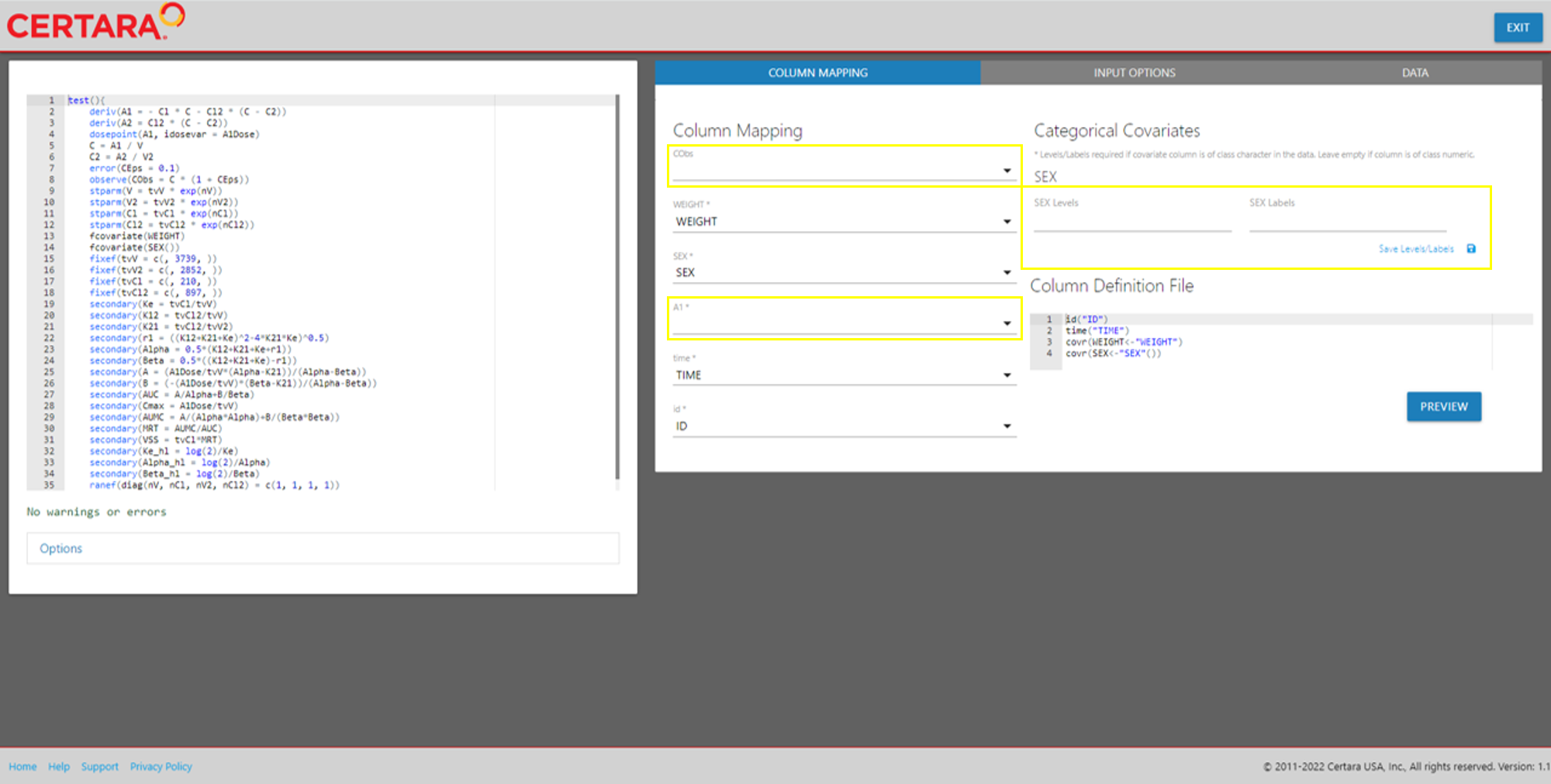
We will need to populate the blank inputs in the above screenshot. Select available columns in the input data from the dropdown inputs to map corresponding model variables. If categorical covariates exist in your model, you will see corresponding “Levels” and “Labels” inputs are displayed in the UI.
If the covariate column is class character in the data, you must specify these inputs and the click “Save Levels/Labels.” Leave empty if the covariate is already encoded as numeric.